Increased Susceptibility and Intrinsic Apoptotic Signaling in Neurons by Induced HDAC3 Expression
- PMID: 34398198
- PMCID: PMC8375002
- DOI: 10.1167/iovs.62.10.14
Increased Susceptibility and Intrinsic Apoptotic Signaling in Neurons by Induced HDAC3 Expression
Abstract
Purpose: Inhibition or targeted deletion of histone deacetylase 3 (HDAC3) is neuroprotective in a variety neurodegenerative conditions, including retinal ganglion cells (RGCs) after acute optic nerve damage. Consistent with this, induced HDAC3 expression in cultured cells shows selective toxicity to neurons. Despite an established role for HDAC3 in neuronal pathology, little is known regarding the mechanism of this pathology.
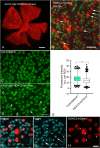
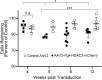
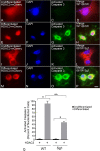
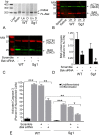
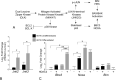
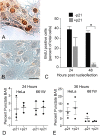
Methods: Induced expression of an HDAC3-mCherry fusion protein in mouse RGCs was accomplished by transduction with AAV2/2-Pgk-HDAC3-mCherry. Increased susceptibility to optic nerve damage in HDAC3-mCherry expressing RGCs was evaluated in transduced mice that received acute optic nerve crush surgery. Expression of HDAC3-FLAG or HDAC3-mCherry was induced by nucleofection or transfection of plasmids into differentiated or undifferentiated 661W tissue culture cells. Immunostaining for cleaved caspase 3, localization of a GFP-BAX fusion protein, and quantitative RT-PCR was used to evaluate HDAC3-induced damage.
Results: Induced expression of exogenous HDAC3 in RGCs by viral-mediated gene transfer resulted in modest levels of cell death but significantly increased the sensitivity of these neurons to axonal damage. Undifferentiated 661W retinal precursor cells were resilient to induced HDAC3 expression, but after differentiation, HDAC3 induced GFP-BAX recruitment to the mitochondria and BAX/BAK dependent activation of caspase 3. This was accompanied by an increase in accumulation of transcripts for the JNK2/3 kinases and the p53-regulated BH3-only gene Bbc3/Puma. Cell cycle arrest of undifferentiated 661W cells did not increase their sensitivity to HDAC3 expression.
Conclusions: Collectively, these results indicate that HDAC3-induced toxicity to neurons is mediated by the intrinsic apoptotic pathway.
Conflict of interest statement
Disclosure:
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

