Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19
- PMID: 34400343
- PMCID: PMC8360988
- DOI: 10.1016/j.cmi.2021.08.009
Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19
Abstract
Objectives: The development of a rapid diagnostic test for viable SARS-CoV-2 is important for infection control. Real-time RT-PCR assays detect non-viable virus, and cell culture differentiates viable virus but it takes several weeks and is labour-intensive. Subgenomic RNAs may reflect replication-competent virus. We therefore evaluated the usefulness of subgenomic RNAs for diagnosing viable SARS-CoV-2 in patients with COVID-19.
Methods: Patients with various severities of confirmed COVID-19 were enrolled at a tertiary hospital between February and December 2020. RT-PCR assay results for genomic and subgenomic RNA of SARS-CoV-2 from nasopharyngeal swab, sputum and saliva specimens were compared with cell culture results.
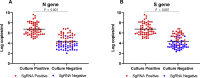
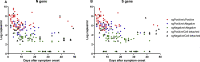
Results: A total 189 specimens from 20 COVID-19 patients were tested in genomic and subgenomic PCR assays and cultured on Vero cells. Of these 189 samples, 62 (33%) gave positive culture results, 93 (49%) negative results and the remaining 34 (18%) indeterminate results. Compared with cell culture results, the sensitivities of genomic RNA and subgenomic RNA of the N and S genes were comparable at 100%, but the specificity of subgenomic RNA (N, 65% and S, 68%) was higher than that of genomic RNA (N, 23% and S, 17%, p < 0.001). The mean durations of positive culture and subgenomic RNA were 11.39 ± 10.34 and 13.75 ± 11.22 days after symptom onset (p 0.437), respectively, while that of genomic RNA was 22.85 ± 11.83 days after symptom onset (p < 0.001).
Discussion: Our comparison of subgenomic RNA detection with symptom duration and SARS-CoV-2 culture positivity provides a significant advancement on the transmissibility-based approach beyond the detection of SARS-CoV-2 genomic RNA, and warrants further studies on the development of better diagnostic strategy.
Keywords: COVID-19; Infective viral shedding; SARS CoV-2; Subgenomic RNA; Virus culture.
Copyright © 2021 European Society of Clinical Microbiology and Infectious Diseases. Published by Elsevier Ltd. All rights reserved.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

