Phase separation of Epstein-Barr virus EBNA2 protein reorganizes chromatin topology for epigenetic regulation
- PMID: 34400762
- PMCID: PMC8368186
- DOI: 10.1038/s42003-021-02501-7
Phase separation of Epstein-Barr virus EBNA2 protein reorganizes chromatin topology for epigenetic regulation
Abstract
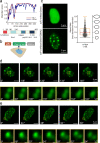
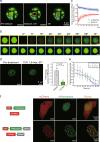
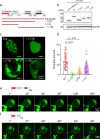
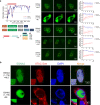
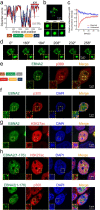
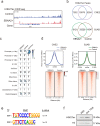
Epstein-Barr virus nuclear antigen 2 (EBNA2) is a transactivator of viral and cellular gene expression, which plays a critical role in the Epstein-Barr virus-associated diseases. It was reported that EBNA2 regulates gene expression by reorganizing chromatin and manipulating epigenetics. Recent studies showed that liquid-liquid phase separation plays an essential role in epigenetic and transcriptional regulation. Here we show that EBNA2 reorganized chromatin topology to form accessible chromatin domains (ACDs) of the host genome by phase separation. The N-terminal region of EBNA2, which is necessary for phase separation, is sufficient to induce ACDs. The C-terminal domain of EBNA2 promotes the acetylation of accessible chromatin regions by recruiting histone acetylase p300 to ACDs. According to these observations, we proposed a model of EBNA2 reorganizing chromatin topology for its acetylation through phase separation to explain the mechanism of EBNA2 hijacking the host genome by controlling its epigenetics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Kempkes B, Ling PD. EBNA2 and its coactivator EBNA-LP. Curr. Top. Microbiol. Immunol. 2015;391:35–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

