The molecular principles of gene regulation by Polycomb repressive complexes
- PMID: 34400841
- PMCID: PMC7612013
- DOI: 10.1038/s41580-021-00398-y
The molecular principles of gene regulation by Polycomb repressive complexes
Abstract
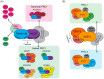
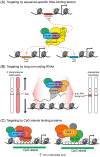
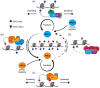
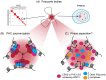
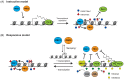
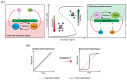
Precise control of gene expression is fundamental to cell function and development. Although ultimately gene expression relies on DNA-binding transcription factors to guide the activity of the transcription machinery to genes, it has also become clear that chromatin and histone post-translational modification have fundamental roles in gene regulation. Polycomb repressive complexes represent a paradigm of chromatin-based gene regulation in animals. The Polycomb repressive system comprises two central protein complexes, Polycomb repressive complex 1 (PRC1) and PRC2, which are essential for normal gene regulation and development. Our early understanding of Polycomb function relied on studies in simple model organisms, but more recently it has become apparent that this system has expanded and diverged in mammals. Detailed studies are now uncovering the molecular mechanisms that enable mammalian PRC1 and PRC2 to identify their target sites in the genome, communicate through feedback mechanisms to create Polycomb chromatin domains and control transcription to regulate gene expression. In this Review, we discuss and contextualize the emerging principles that define how this fascinating chromatin-based system regulates gene expression in mammals.
© 2021. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017;171:34–57. - PubMed
-
- de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. - PubMed
-
- Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

