Sclerosing Sialadenitis Is Associated With Salivary Gland Hypofunction and a Unique Gene Expression Profile in Sjögren's Syndrome
- PMID: 34400910
- PMCID: PMC8363566
- DOI: 10.3389/fimmu.2021.699722
Sclerosing Sialadenitis Is Associated With Salivary Gland Hypofunction and a Unique Gene Expression Profile in Sjögren's Syndrome
Abstract
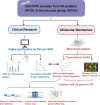
Purpose: To develop a novel method to quantify the amount of fibrosis in the salivary gland and to investigate the relationship between fibrosis and specific symptoms associated with Sjögren's syndrome (SS) using this method.
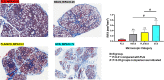
Materials and methods: Paraffin-embedded labial salivary gland (LSG) slides from 20 female SS patients and their clinical and LSG pathology data were obtained from the Sjögren's International Collaborative Clinical Alliance. Relative interstitial fibrosis area (RIFA) in Masson's trichrome-stained LSG sections was quantified from digitally scanned slides and used for correlation analysis. Gene expression levels were assessed by microarray analysis. Core promoter accessibility for RIFA-correlated genes was determined using DNase I hypersensitive sites sequencing analysis.
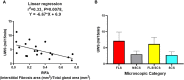
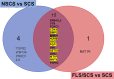
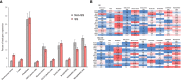
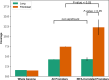
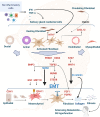
Results: RIFA was significantly correlated with unstimulated whole saliva flow rate in SS patients. Sixteen genes were significantly and positively correlated with RIFA. In a separate analysis, a group of differentially expressed genes was identified by comparing severe and moderate fibrosis groups. This combined set of genes was distinct from differentially expressed genes identified in lung epithelium from idiopathic pulmonary fibrosis patients compared with controls. Single-cell RNA sequencing analysis of salivary glands suggested most of the RIFA-correlated genes are expressed by fibroblasts in the gland and are in a permissive chromatin state.
Conclusion: RIFA quantification is a novel method for assessing interstitial fibrosis and the impact of fibrosis on SS symptoms. Loss of gland function may be associated with salivary gland fibrosis, which is likely to be driven by a unique set of genes that are mainly expressed by fibroblasts.
Trial registration: ClinicalTrials.gov NCT02327884.
Keywords: Sjögren’s syndrome; salivary gland hypofunction; salivary gland interstitial fibrosis; sclerosing sialadenitis; transcriptomic gene expression profile.
Copyright © 2021 Yin, Pranzatelli, French, Zhang, Warner, Chiorini and NIDCD/NIDCR Genomics and Computational Biology Core.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Associations between salivary gland histopathologic diagnoses and phenotypic features of Sjögren's syndrome among 1,726 registry participants.Arthritis Rheum. 2011 Jul;63(7):2021-30. doi: 10.1002/art.30381. Arthritis Rheum. 2011. PMID: 21480190 Free PMC article.
-
[Biopsy of the minor salivary glands in the differential diagnosis of Sjögren's syndrome and chronic sialadenitis].Ter Arkh. 1988;60(4):38-9. Ter Arkh. 1988. PMID: 3394094 Russian.
-
Ultrasound-guided core needle biopsy compared with open biopsy: a new diagnostic approach to salivary gland enlargement in Sjögren's syndrome?Rheumatology (Oxford). 2021 Mar 2;60(3):1282-1290. doi: 10.1093/rheumatology/keaa441. Rheumatology (Oxford). 2021. PMID: 32940706
-
Immune and non-immune mediators in the fibrosis pathogenesis of salivary gland in Sjögren's syndrome.Front Immunol. 2024 Oct 14;15:1421436. doi: 10.3389/fimmu.2024.1421436. eCollection 2024. Front Immunol. 2024. PMID: 39469708 Free PMC article. Review.
-
[Lymphoproliferative disorders in Sjögren's syndrome].Otolaryngol Pol. 2005;59(4):559-64. Otolaryngol Pol. 2005. PMID: 16273862 Review. Polish.
Cited by
-
Temporal evolution of fibroblast responses following salivary gland ductal ligation injury.Front Dent Med. 2025 May 1;6:1581376. doi: 10.3389/fdmed.2025.1581376. eCollection 2025. Front Dent Med. 2025. PMID: 40375832 Free PMC article.
-
Lysosome-Associated Membrane Protein 3 Induces Lysosome-Dependent Cell Death by Impairing Autophagic Caspase 8 Degradation in the Salivary Glands of Individuals With Sjögren's Disease.Arthritis Rheumatol. 2023 Sep;75(9):1586-1598. doi: 10.1002/art.42540. Epub 2023 Jul 27. Arthritis Rheumatol. 2023. PMID: 37096570 Free PMC article.
-
Hyperosmolar environment and salivary gland epithelial cells increase extra-cellular matrix remodeling and lymphocytic infiltration in Sjögren's syndrome.Clin Exp Immunol. 2023 Apr 7;212(1):39-51. doi: 10.1093/cei/uxad020. Clin Exp Immunol. 2023. PMID: 36759947 Free PMC article.
-
Salivary gland LAMP3 mRNA expression is a possible predictive marker in the response to hydroxychloroquine in Sjögren's disease.PLoS One. 2023 Feb 23;18(2):e0282227. doi: 10.1371/journal.pone.0282227. eCollection 2023. PLoS One. 2023. PMID: 36821638 Free PMC article.
-
TScan-II: A genome-scale platform for the de novo identification of CD4+ T cell epitopes.Cell. 2023 Dec 7;186(25):5569-5586.e21. doi: 10.1016/j.cell.2023.10.024. Epub 2023 Nov 27. Cell. 2023. PMID: 38016469 Free PMC article.
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

