Identification of natural compounds as SARS-CoV-2 entry inhibitors by molecular docking-based virtual screening with bio-layer interferometry
- PMID: 34403732
- PMCID: PMC8364251
- DOI: 10.1016/j.phrs.2021.105820
Identification of natural compounds as SARS-CoV-2 entry inhibitors by molecular docking-based virtual screening with bio-layer interferometry
Abstract
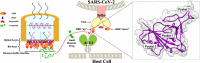
Coronavirus Disease 2019 (COVID-19) is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which enter the host cells through the interaction between its receptor binding domain (RBD) of spike glycoprotein with angiotensin-converting enzyme 2 (ACE2) receptor on the plasma membrane of host cell. Neutralizing antibodies and peptide binders of RBD can block viral infection, however, the concern of accessibility and affordability of viral infection inhibitors has been raised. Here, we report the identification of natural compounds as potential SARS-CoV-2 entry inhibitors using the molecular docking-based virtual screening coupled with bilayer interferometry (BLI). From a library of 1871 natural compounds, epigallocatechin gallate (EGCG), 20(R)-ginsenoside Rg3 (RRg3), 20(S)-ginsenoside Rg3 (SRg3), isobavachalcone (Ibvc), isochlorogenic A (IscA) and bakuchiol (Bkc) effectively inhibited pseudovirus entry at concentrations up to 100 μM. Among these compounds, four compounds, EGCG, Ibvc, salvianolic acid A (SalA), and isoliensinine (Isl), were effective in inhibiting SARS-CoV-2-induced cytopathic effect and plaque formation in Vero E6 cells. The EGCG was further validated with no observable animal toxicity and certain antiviral effect against SARS-CoV-2 pseudovirus mutants (D614G, N501Y, N439K & Y453F). Interestingly, EGCG, Bkc and Ibvc bind to ACE2 receptor in BLI assay, suggesting a dual binding to RBD and ACE2. Current findings shed some insight into identifications and validations of SARS-CoV-2 entry inhibitors from natural compounds.
Keywords: Bio-layer interferometry; Competitive binding; Molecular docking; Molecular dynamics simulation; Natural compounds; SARS-CoV-2.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions.mBio. 2021 Mar 30;12(2):e03681-20. doi: 10.1128/mBio.03681-20. mBio. 2021. PMID: 33785634 Free PMC article.
-
Identification of nitrile-containing isoquinoline-related natural product derivatives as coronavirus entry inhibitors in silico and in vitro.Biomed Pharmacother. 2024 Nov;180:117517. doi: 10.1016/j.biopha.2024.117517. Epub 2024 Oct 1. Biomed Pharmacother. 2024. PMID: 39357326
-
Corilagin prevents SARS-CoV-2 infection by targeting RBD-ACE2 binding.Phytomedicine. 2021 Jul;87:153591. doi: 10.1016/j.phymed.2021.153591. Epub 2021 May 5. Phytomedicine. 2021. PMID: 34029937 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Screening S protein - ACE2 blockers from natural products: Strategies and advances in the discovery of potential inhibitors of COVID-19.Eur J Med Chem. 2021 Dec 15;226:113857. doi: 10.1016/j.ejmech.2021.113857. Epub 2021 Oct 4. Eur J Med Chem. 2021. PMID: 34628234 Free PMC article. Review.
Cited by
-
Roles of ginsenosides in sepsis.J Ginseng Res. 2023 Jan;47(1):1-8. doi: 10.1016/j.jgr.2022.05.004. Epub 2022 May 7. J Ginseng Res. 2023. PMID: 36644389 Free PMC article. Review.
-
Potential of green tea EGCG in neutralizing SARS-CoV-2 Omicron variant with greater tropism toward the upper respiratory tract.Trends Food Sci Technol. 2023 Feb;132:40-53. doi: 10.1016/j.tifs.2022.12.012. Epub 2022 Dec 28. Trends Food Sci Technol. 2023. PMID: 36594074 Free PMC article. Review.
-
Structure-based docking, pharmacokinetic evaluation, and molecular dynamics-guided evaluation of traditional formulation against SARS-CoV-2 spike protein receptor bind domain and ACE2 receptor complex.Chem Zvesti. 2022;76(2):1063-1083. doi: 10.1007/s11696-021-01917-z. Epub 2021 Oct 18. Chem Zvesti. 2022. PMID: 34690412 Free PMC article.
-
Colloidal Aggregation Confounds Cell-Based Covid-19 Antiviral Screens.J Med Chem. 2024 Jun 27;67(12):10263-10274. doi: 10.1021/acs.jmedchem.4c00597. Epub 2024 Jun 12. J Med Chem. 2024. PMID: 38864383 Free PMC article.
-
Exploring new catechin derivatives as SARS-CoV-2 Mpro inhibitors from tea by molecular networking, surface plasma resonance, enzyme inhibition, induced fit docking, and metadynamics simulations.Comput Biol Med. 2022 Dec;151(Pt A):106288. doi: 10.1016/j.compbiomed.2022.106288. Epub 2022 Nov 12. Comput Biol Med. 2022. PMID: 36401970 Free PMC article.
References
-
- Brian D.A., Baric R.S. In: Coronavirus Replication and Reverse Genetics. Enjuanes L., editor. Springer; Berlin Heidelberg: 2005. Coronavirus genome structure and replication; pp. 1–30.
-
- Wang K., Chen W., Zhang Z., Deng Y., Lian J.-Q., Du P., Wei D., Zhang Y., Sun X.-X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.-J., Chen R., Zhang H., Wang B., Zhu Y.-M., Nan G., Jiang J.-L., Li L., Wu J., Lin P., Huang W., Xie L., Zheng Z.-H., Zhang K., Miao J.-L., Cui H.-Y., Huang M., Zhang J., Fu L., Yang X.-M., Zhao Z., Sun S., Gu H., Wang Z., Wang C.-F., Lu Y., Liu Y.-Y., Wang Q.-Y., Bian H., Zhu P., Chen Z.-N. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020;5(1):283. - PMC - PubMed
-
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Österlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

