Dynamic landscape mapping of humoral immunity to SARS-CoV-2 identifies non-structural protein antibodies associated with the survival of critical COVID-19 patients
- PMID: 34404759
- PMCID: PMC8368053
- DOI: 10.1038/s41392-021-00718-w
Dynamic landscape mapping of humoral immunity to SARS-CoV-2 identifies non-structural protein antibodies associated with the survival of critical COVID-19 patients
Abstract
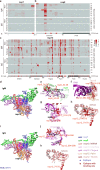
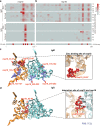
A comprehensive analysis of the humoral immune response to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is essential in understanding COVID-19 pathogenesis and developing antibody-based diagnostics and therapy. In this work, we performed a longitudinal analysis of antibody responses to SARS-CoV-2 proteins in 104 serum samples from 49 critical COVID-19 patients using a peptide-based SARS-CoV-2 proteome microarray. Our data show that the binding epitopes of IgM and IgG antibodies differ across SARS-CoV-2 proteins and even within the same protein. Moreover, most IgM and IgG epitopes are located within nonstructural proteins (nsps), which are critical in inactivating the host's innate immune response and enabling SARS-CoV-2 replication, transcription, and polyprotein processing. IgM antibodies are associated with a good prognosis and target nsp3 and nsp5 proteases, whereas IgG antibodies are associated with high mortality and target structural proteins (Nucleocapsid, Spike, ORF3a). The epitopes targeted by antibodies in patients with a high mortality rate were further validated using an independent serum cohort (n = 56) and using global correlation mapping analysis with the clinical variables that are associated with COVID-19 severity. Our data provide fundamental insight into humoral immunity during SARS-CoV-2 infection. SARS-CoV-2 immunogenic epitopes identified in this work could also help direct antibody-based COVID-19 treatment and triage patients.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

