Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern
- PMID: 34404775
- PMCID: PMC8371089
- DOI: 10.1038/s41467-021-25167-5
Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern
Abstract
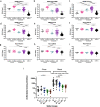
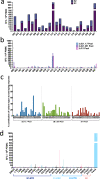
The extent to which immune responses to natural infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and immunization with vaccines protect against variants of concern (VOC) is of increasing importance. Accordingly, here we analyse antibodies and T cells of a recently vaccinated, UK cohort, alongside those recovering from natural infection in early 2020. We show that neutralization of the VOC compared to a reference isolate of the original circulating lineage, B, is reduced: more profoundly against B.1.351 than for B.1.1.7, and in responses to infection or a single dose of vaccine than to a second dose of vaccine. Importantly, high magnitude T cell responses are generated after two vaccine doses, with the majority of the T cell response directed against epitopes that are conserved between the prototype isolate B and the VOC. Vaccination is required to generate high potency immune responses to protect against these and other emergent variants.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- World Health Organization. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants (2021).
-
- GISAID. GISAID Initiative. https://www.gisaid.org/ (2021).
-
- Verkhivker, G. M., Agajanian, S., Oztas, D. & Gupta, G. Computational analysis of protein stability and allosteric interaction networks in distinct conformational forms of the SARS-CoV-2 Spike D614G mutant: reconciling functional mechanisms through allosteric model of spike regulation. J. Biomol. Struct. Dyn.10.1080/07391102.2021.1933594 (2021). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- MC_PC_20060/MRC_/Medical Research Council/United Kingdom
- MR/V028448/1/MRC_/Medical Research Council/United Kingdom
- MR/V027859/1/MRC_/Medical Research Council/United Kingdom
- MR/L006588/1/MRC_/Medical Research Council/United Kingdom
- RE/18/3/34214/BHF_/British Heart Foundation/United Kingdom
- NIHR200907/DH_/Department of Health/United Kingdom
- 211153/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- NIHR300791/DH_/Department of Health/United Kingdom
- 109965/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 216417/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- WT109965MA /WT_/Wellcome Trust/United Kingdom
- 204721/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

