Genetic diversity analysis of Korean peanut germplasm using 48 K SNPs 'Axiom_Arachis' Array and its application for cultivar differentiation
- PMID: 34404839
- PMCID: PMC8371136
- DOI: 10.1038/s41598-021-96074-4
Genetic diversity analysis of Korean peanut germplasm using 48 K SNPs 'Axiom_Arachis' Array and its application for cultivar differentiation
Abstract
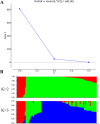
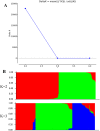
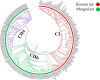
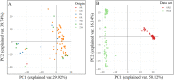
Cultivated peanut (Arachis hypogaea) is one of the important legume oilseed crops. Cultivated peanut has a narrow genetic base. Therefore, it is necessary to widen its genetic base and diversity for additional use. The objective of the present study was to assess the genetic diversity and population structure of 96 peanut genotypes with 9478 high-resolution SNPs identified from a 48 K 'Axiom_Arachis' SNP array. Korean set genotypes were also compared with a mini-core of US genotypes. These sets of genotypes were used for genetic diversity analysis. Model-based structure analysis at K = 2 indicated the presence of two subpopulations in both sets of genotypes. Phylogenetic and PCA analysis clustered these genotypes into two major groups. However, clear genotype distribution was not observed for categories of subspecies, botanical variety, or origin. The analysis also revealed that current Korean genetic resources lacked variability compared to US mini-core genotypes. These results suggest that Korean genetic resources need to be expanded by creating new allele combinations and widening the genetic pool to offer new genetic variations for Korean peanut improvement programs. High-quality SNP data generated in this study could be used for identifying varietal contaminant, QTL, and genes associated with desirable traits by performing mapping, genome-wide association studies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

