How RNA modifications regulate the antiviral response
- PMID: 34405413
- PMCID: PMC8616813
- DOI: 10.1111/imr.13020
How RNA modifications regulate the antiviral response
Abstract
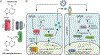
Induction of the antiviral innate immune response is highly regulated at the RNA level, particularly by RNA modifications. Recent discoveries have revealed how RNA modifications play key roles in cellular surveillance of nucleic acids and in controlling gene expression in response to viral infection. These modifications have emerged as being essential for a functional antiviral response and maintaining cellular homeostasis. In this review, we will highlight these and other discoveries that describe how the antiviral response is controlled by modifications to both viral and cellular RNA, focusing on how mRNA cap modifications, N6-methyladenosine, and RNA editing all contribute to coordinating an efficient response that properly controls viral infection.
Keywords: N6-methyladenosine; RNA editing; adenosine deaminases acting on RNA; cap modification; innate immunity; interferon; pattern recognition receptors.
© 2021 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Conflict of interest statement
Conflict of Interest Statement
M.G.T, M.T.S., and S.M.H. declare no conflicts of interest with the contents of this article.
Figures


References
-
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

