Saliva is Comparable to Nasopharyngeal Swabs for Molecular Detection of SARS-CoV-2
- PMID: 34406838
- PMCID: PMC8552668
- DOI: 10.1128/Spectrum.00162-21
Saliva is Comparable to Nasopharyngeal Swabs for Molecular Detection of SARS-CoV-2
Abstract
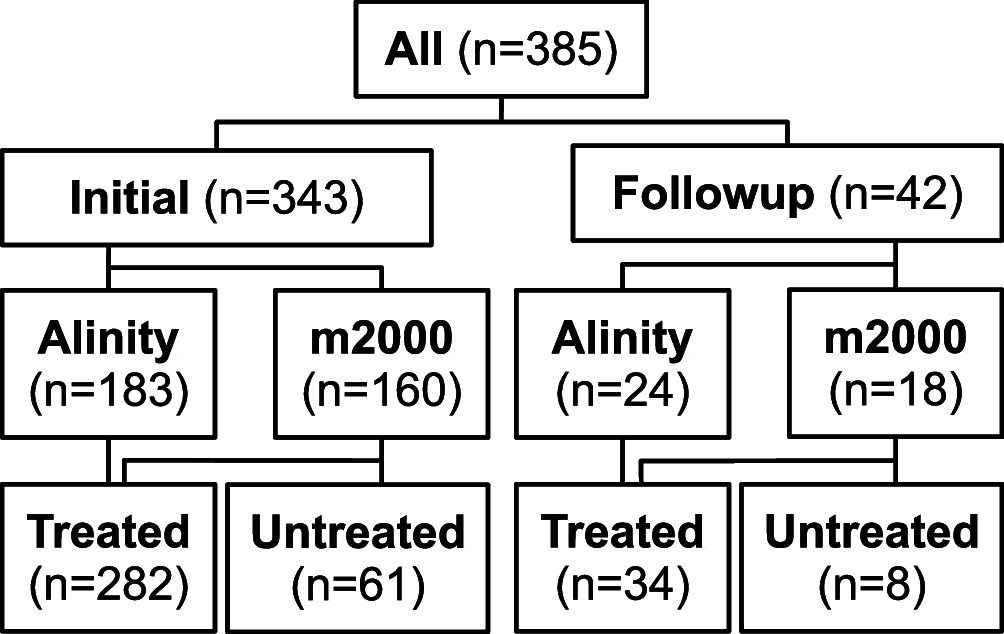
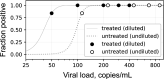
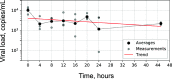
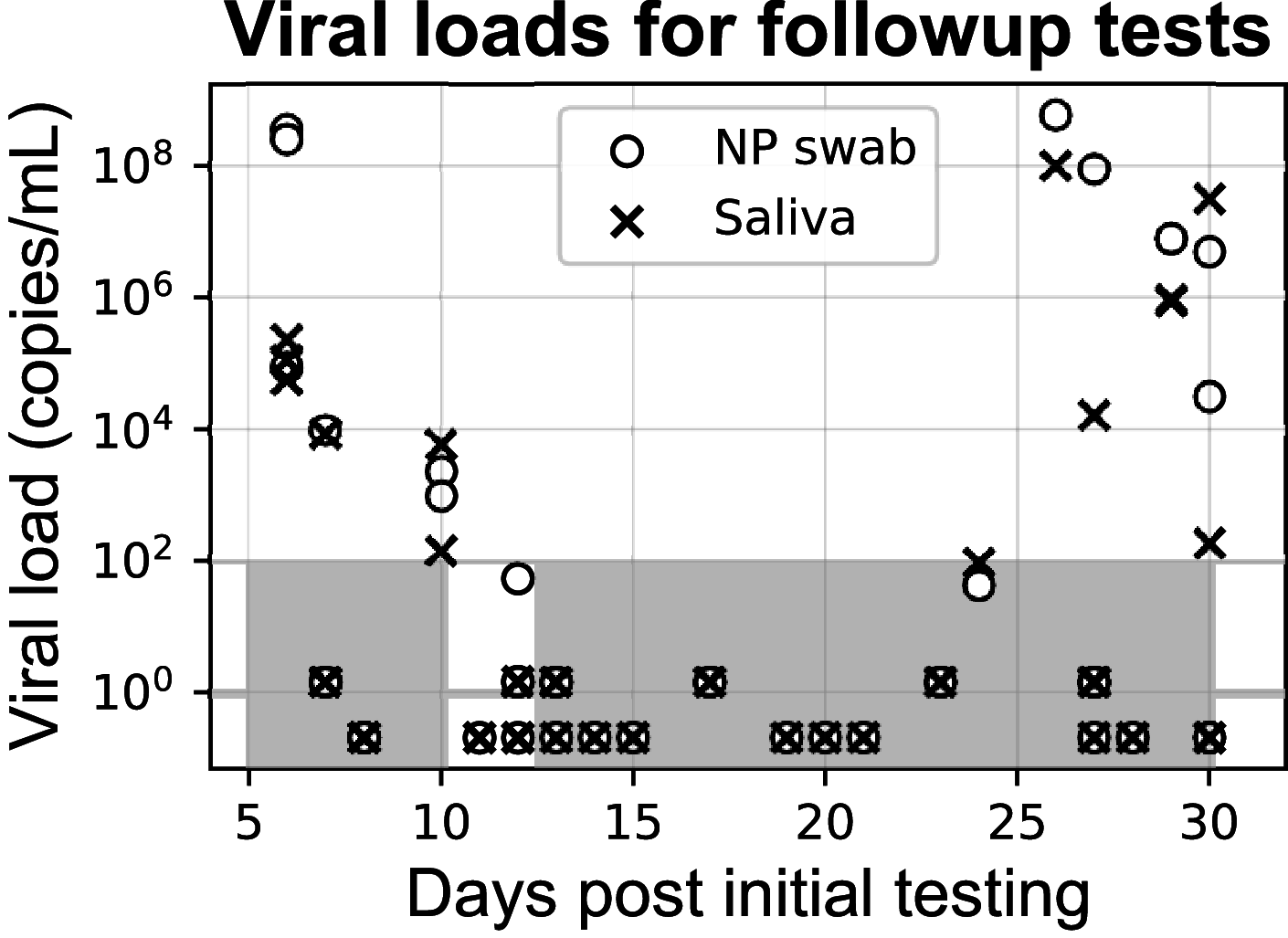
The continued need for molecular testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the potential for self-collected saliva as an alternative to nasopharyngeal (NP) swabs for sample acquisition led us to compare saliva to NP swabs in an outpatient setting without restrictions to avoid food, drink, smoking, or tooth-brushing. A total of 385 pairs of NP and saliva specimens were obtained, the majority from individuals presenting for initial evaluation, and were tested on two high-sensitivity reverse transcriptase PCR (RT-PCR) platforms, the Abbott m2000 and Abbott Alinity m (both with limits of detection [LoD] of 100 copies of viral RNA/ml). Concordance between saliva and NP swabs was excellent overall (Cohen's κ = 0.93) for both initial and follow-up testing, for both platforms, and for specimens treated with guanidinium transport medium as preservative as well as for untreated saliva (κ = 0.88 to 0.95). Viral loads were on average 16× higher in NP specimens than saliva specimens, suggesting that only the relatively small fraction of outpatients (∼8% in this study) who present with very low viral loads (<1,600 copies/ml from NP swabs) would be missed by testing saliva instead of NP swabs when using sensitive testing platforms. Special attention was necessary to ensure leak-resistant specimen collection and transport. The advantages of self-collection of saliva, without behavioral restrictions, will likely outweigh a minor potential decrease in clinical sensitivity in individuals less likely to pose an infectious risk to others for many real-world scenarios, especially for initial testing. IMPORTANCE In general, the most accurate COVID-19 testing is hands-on and uncomfortable, requiring trained staff and a "brain-tickling" nasopharyngeal swab. Saliva would be much easier on both fronts, since patients could collect it themselves, and it is after all just spit. However, despite much interest, it remains unclear how well saliva performs in real-world settings when just using it in place of an NP swab without elaborate or cumbersome restrictions about not eating/drinking before testing, etc. Also, almost all studies of COVID-19 testing, whether of NP swabs, saliva, or otherwise, have been restricted to reporting results in the abstruse units of "CT values," which only mean something in the context of a specific assay and testing platform. Here, we compared saliva versus NP swabs in a real-world setting without restriction and report all results in natural units-the amount of virus being shed-showing that saliva is essentially just as good as NP swabs.
Keywords: COVID-19; NP swab; SARS-CoV-2; limit of detection; saliva.
Figures

Comment in
-
Kappa Values in Testing the Concordance: Comments on a Recent Article about Nasopharyngeal Swabs for SARS-CoV-2 Detection.Microbiol Spectr. 2022 Dec 21;10(6):e0158222. doi: 10.1128/spectrum.01582-22. Epub 2022 Nov 1. Microbiol Spectr. 2022. PMID: 36318017 Free PMC article. No abstract available.
-
Reply to Li et al., "Kappa Values in Testing the Concordance: Comments on a Recent Article about Nasopharyngeal Swabs for SARS-CoV-2 Detection".Microbiol Spectr. 2022 Dec 21;10(6):e0341822. doi: 10.1128/spectrum.03418-22. Epub 2022 Nov 1. Microbiol Spectr. 2022. PMID: 36318026 Free PMC article. No abstract available.
Similar articles
-
Comparison of SARS-CoV-2 PCR-Based Detection Using Saliva or Nasopharyngeal Swab Specimens in Asymptomatic Populations.Microbiol Spectr. 2021 Sep 3;9(1):e0006221. doi: 10.1128/Spectrum.00062-21. Epub 2021 Aug 25. Microbiol Spectr. 2021. PMID: 34431689 Free PMC article.
-
Diagnostic Performance of Self-Collected Saliva Versus Nasopharyngeal Swab for the Molecular Detection of SARS-CoV-2 in the Clinical Setting.Microbiol Spectr. 2021 Dec 22;9(3):e0046821. doi: 10.1128/Spectrum.00468-21. Epub 2021 Nov 3. Microbiol Spectr. 2021. PMID: 34730436 Free PMC article.
-
A Multiplex One-Step RT-qPCR Protocol to Detect SARS-CoV-2 in NP/OP Swabs and Saliva.Curr Protoc. 2021 May;1(5):e145. doi: 10.1002/cpz1.145. Curr Protoc. 2021. PMID: 34004070 Free PMC article.
-
Screening for SARS-CoV-2 by RT-PCR: Saliva or nasopharyngeal swab? Rapid review and meta-analysis.PLoS One. 2021 Jun 10;16(6):e0253007. doi: 10.1371/journal.pone.0253007. eCollection 2021. PLoS One. 2021. PMID: 34111196 Free PMC article.
-
Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis.Lancet Infect Dis. 2021 Sep;21(9):1233-1245. doi: 10.1016/S1473-3099(21)00146-8. Epub 2021 Apr 12. Lancet Infect Dis. 2021. PMID: 33857405 Free PMC article.
Cited by
-
Advancements in SARS-CoV-2 Testing: Enhancing Accessibility through Machine Learning-Enhanced Biosensors.Micromachines (Basel). 2023 Jul 28;14(8):1518. doi: 10.3390/mi14081518. Micromachines (Basel). 2023. PMID: 37630054 Free PMC article. Review.
-
Salivary SARS-CoV-2 RNA for diagnosis of COVID-19 patients: a systematic revisew and meta-analysis of diagnostic accuracy.Jpn Dent Sci Rev. 2023 Jun 21;59:219-38. doi: 10.1016/j.jdsr.2023.06.004. Online ahead of print. Jpn Dent Sci Rev. 2023. PMID: 37360001 Free PMC article. Review.
-
Saliva diagnostics: Salivaomics, saliva exosomics, and saliva liquid biopsy.J Am Dent Assoc. 2023 Aug;154(8):696-704. doi: 10.1016/j.adaj.2023.05.006. J Am Dent Assoc. 2023. PMID: 37500232 Free PMC article. Review.
-
Kappa Values in Testing the Concordance: Comments on a Recent Article about Nasopharyngeal Swabs for SARS-CoV-2 Detection.Microbiol Spectr. 2022 Dec 21;10(6):e0158222. doi: 10.1128/spectrum.01582-22. Epub 2022 Nov 1. Microbiol Spectr. 2022. PMID: 36318017 Free PMC article. No abstract available.
-
Quantitative SARS-CoV-2 viral-load curves in paired saliva and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection.medRxiv [Preprint]. 2021 Aug 26:2021.04.02.21254771. doi: 10.1101/2021.04.02.21254771. medRxiv. 2021. Update in: J Clin Microbiol. 2022 Feb 16;60(2):e0178521. doi: 10.1128/JCM.01785-21. PMID: 33851180 Free PMC article. Updated. Preprint.
References
-
- Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. 2020. Comparison of Abbott ID Now, Diasorin Simplexa, and CDC FDA emergency use authorization methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol 58:e00760-20. doi:10.1128/JCM.00760-20. - DOI - PMC - PubMed
-
- McCormick-Baw C, Morgan K, Gaffney D, Cazares Y, Jaworski K, Byrd A, Molberg K, Cavuoti D. 2020. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol 58:e01109-20. doi:10.1128/JCM.01109-20. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

