Plastome variation and phylogeny of Taxillus (Loranthaceae)
- PMID: 34407123
- PMCID: PMC8372910
- DOI: 10.1371/journal.pone.0256345
Plastome variation and phylogeny of Taxillus (Loranthaceae)
Abstract
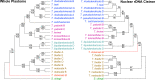
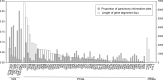
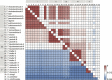
Several molecular phylogenetic studies of the mistletoe family Loranthaceae have been published such that now the general pattern of relationships among the genera and their biogeographic histories are understood. Less is known about species relationships in the larger (> 10 species) genera. This study examines the taxonomically difficult genus Taxillus composed of 35-40 Asian species. The goal was to explore the genetic diversity present in Taxillus plastomes, locate genetically variable hotspots, and test these for their utility as potential DNA barcodes. Using genome skimming, complete plastomes, as well as nuclear and mitochondrial rDNA sequences, were newly generated for eight species. The plastome sequences were used in conjunction with seven publicly available Taxillus sequences and three sequences of Scurrula, a close generic relative. The Taxillus plastomes ranged from 121 to 123 kbp and encoded 90-93 plastid genes. In addition to all of the NADH dehydrogenase complex genes, four ribosomal genes, infA and four intron-containing tRNA genes were lost or pseudogenized in all of the Taxillus and Scurrula plastomes. The topologies of the plastome, mitochondrial rDNA and nuclear rDNA trees were generally congruent, though with discordance at the position of T. chinensis. Several variable regions in the plastomes were identified that have sufficient numbers of parsimony informative sites as to recover the major clades seen in the complete plastome tree. Instead of generating complete plastome sequences, our study showed that accD alone or the concatenation of accD and rbcL can be used in future studies to facilitate identification of Taxillus samples and to generate a molecular phylogeny with robust sampling within the genus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Su H-J, Hu J-M, Anderson FE, Der JP, Nickrent DL. Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae. Taxon. 2015;64(3):491–506. doi: 10.12705/643.2 - DOI
-
- Nickrent DL. Parasitic angiosperms: How often and how many? Taxon. 2020;69(1):5–27. doi: 10.1002/tax.12195 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

