Selective autophagy
- PMID: 34407274
- PMCID: PMC8486182
- DOI: 10.1111/cas.15112
Selective autophagy
Abstract
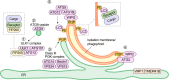
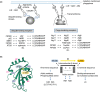
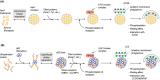
While starvation-induced autophagy is thought to randomly degrade cellular components, under certain circumstances autophagy selectively recognizes, sequesters, and degrades specific targets via autophagosomes. This process is called selective autophagy, and it contributes to cellular homeostasis by degrading specific soluble proteins, supramolecular complexes, liquid-liquid phase-separated droplets, abnormal or excess organelles, and pathogenic invasive bacteria. This means that autophagy, like the ubiquitin-proteasome system, strictly regulates diverse cellular functions through its selectivity. In this short review, we focus on the mechanism of "selective" autophagy, which is rapidly being elucidated.
Keywords: ATG8-family proteins; autophagy; liquid-liquid phase separation; selective autophagy; selective autophagy receptors.
© 2021 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
The authors have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

