Syntax-based transfer learning for the task of biomedical relation extraction
- PMID: 34407869
- PMCID: PMC8371836
- DOI: 10.1186/s13326-021-00248-y
Syntax-based transfer learning for the task of biomedical relation extraction
Abstract
Background: Transfer learning aims at enhancing machine learning performance on a problem by reusing labeled data originally designed for a related, but distinct problem. In particular, domain adaptation consists for a specific task, in reusing training data developedfor the same task but a distinct domain. This is particularly relevant to the applications of deep learning in Natural Language Processing, because they usually require large annotated corpora that may not exist for the targeted domain, but exist for side domains.
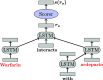
Results: In this paper, we experiment with transfer learning for the task of relation extraction from biomedical texts, using the TreeLSTM model. We empirically show the impact of TreeLSTM alone and with domain adaptation by obtaining better performances than the state of the art on two biomedical relation extraction tasks and equal performances for two others, for which little annotated data are available. Furthermore, we propose an analysis of the role that syntactic features may play in transfer learning for relation extraction.
Conclusion: Given the difficulty to manually annotate corpora in the biomedical domain, the proposed transfer learning method offers a promising alternative to achieve good relation extraction performances for domains associated with scarce resources. Also, our analysis illustrates the importance that syntax plays in transfer learning, underlying the importance in this domain to privilege approaches that embed syntactic features.
Keywords: Biomedical relation extraction; Deep learning; Transfer learning.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Weiss KR, Khoshgoftaar TM, Wang D. A survey of transfer learning. J Big Data. 2016;3:9.
-
- Zeng D, Liu K, Lai S, Zhou G, Zhao J. Proceedings of COLING 2014, the 25th International Conference on Computational Linguistics: Technical Papers. Dublin: Dublin City University and Association for Computational Linguistics; 2014. Relation classification via convolutional deep neural network.
-
- Mintz M, Bills S, Snow R, Jurafsky D. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP. Suntec: Association for Computational Linguistics; 2009. Distant supervision for relation extraction without labeled data.
-
- van Mulligen EM, Fourrier-Réglat A, Gurwitz D, Molokhia M, Nieto A, Trifirò G, Kors JA, Furlong LI. The EU-ADR corpus: Annotated drugs, diseases, targets, and their relationships. J Biomed Inform. 2012;45(5):879–84. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

