The structure and flexibility analysis of the Arabidopsis synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
- PMID: 34408000
- PMCID: PMC8380656
- DOI: 10.26508/lsa.202101152
The structure and flexibility analysis of the Arabidopsis synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Abstract
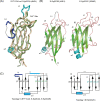
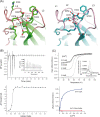
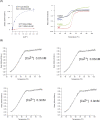
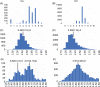
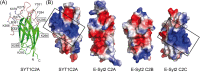
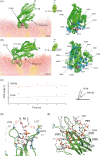
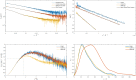
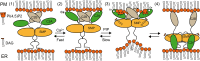
Non-vesicular lipid transfer at ER and plasma membrane (PM) contact sites (CS) is crucial for the maintenance of membrane lipid homeostasis. Extended synaptotagmins (E-Syts) play a central role in this process as they act as molecular tethers of ER and PM and as lipid transfer proteins between these organelles. E-Syts are proteins constitutively anchored to the ER through an N-terminal hydrophobic segment and bind the PM via a variable number of C-terminal C2 domains. Synaptotagmins (SYTs) are the plant orthologous of E-Syts and regulate the ER-PM communication in response to abiotic stress. Combining different structural and biochemical techniques, we demonstrate that the binding of SYT1 to lipids occurs through a Ca2+-dependent lipid-binding site and by a site for phosphorylated forms of phosphatidylinositol, thus integrating two different molecular signals in response to stress. In addition, we show that SYT1 displays three highly flexible hinge points that provide conformational freedom to facilitate lipid extraction, protein loading, and subsequent transfer between PM and ER.
© 2021 Benavente et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. 10.1107/S0907444909052925 - DOI - PMC - PubMed
-
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. (2014) Swiss-model: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252–W258. 10.1093/nar/gku340 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
