Adipose stem cell niche reprograms the colorectal cancer stem cell metastatic machinery
- PMID: 34408135
- PMCID: PMC8373975
- DOI: 10.1038/s41467-021-25333-9
Adipose stem cell niche reprograms the colorectal cancer stem cell metastatic machinery
Abstract
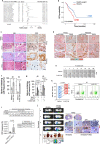
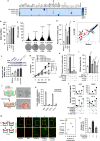
Obesity is a strong risk factor for cancer progression, posing obesity-related cancer as one of the leading causes of death. Nevertheless, the molecular mechanisms that endow cancer cells with metastatic properties in patients affected by obesity remain unexplored.Here, we show that IL-6 and HGF, secreted by tumor neighboring visceral adipose stromal cells (V-ASCs), expand the metastatic colorectal (CR) cancer cell compartment (CD44v6 + ), which in turn secretes neurotrophins such as NGF and NT-3, and recruits adipose stem cells within tumor mass. Visceral adipose-derived factors promote vasculogenesis and the onset of metastatic dissemination by activation of STAT3, which inhibits miR-200a and enhances ZEB2 expression, effectively reprogramming CRC cells into a highly metastatic phenotype. Notably, obesity-associated tumor microenvironment provokes a transition in the transcriptomic expression profile of cells derived from the epithelial consensus molecular subtype (CMS2) CRC patients towards a mesenchymal subtype (CMS4). STAT3 pathway inhibition reduces ZEB2 expression and abrogates the metastatic growth sustained by adipose-released proteins. Together, our data suggest that targeting adipose factors in colorectal cancer patients with obesity may represent a therapeutic strategy for preventing metastatic disease.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Bluher, M. Obesity: global epidemiology and pathogenesis. Nature reviews Endocrinology.15, 288–298 (2019). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

