Investigation of Potential Molecular Biomarkers for Diagnosis and Prognosis of AFP-Negative HCC
- PMID: 34408477
- PMCID: PMC8364386
- DOI: 10.2147/IJGM.S323868
Investigation of Potential Molecular Biomarkers for Diagnosis and Prognosis of AFP-Negative HCC
Abstract
Background: Alpha-fetoprotein (AFP) is the most important diagnostic and prognostic index of hepatocellular carcinoma (HCC). AFP-positive HCC can be easily diagnosed based on the serum AFP level and typical imaging features, but a number of HCC patients are negative (AFP < 20 ng/mL) for AFP. Therefore, it is necessary to develop novel diagnostic and prognostic biomarkers for AFP-negative HCC.
Methods: RNA data from TCGA and differential expression of lncRNAs, miRNAs, and mRNAs were downloaded to analyze the differential RNA expression patterns between AFP-negative HCC tissues and normal tissues. A lncRNA-miRNA-mRNA ceRNA regulatory network was constructed to elucidate the interaction mechanism of RNAs. Functional enrichment analysis of these DEmRNAs was performed to indirectly reveal the mechanism of action of lncRNAs. A PPI network was built using STRING, and the hub genes were identified with Cytoscape. The diagnostic value of hub genes was assessed with receiver operating characteristic (ROC) analysis. And the prognostic value of RNAs in the ceRNA was estimated with Kaplan-Meier curve analysis.
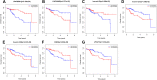
Results: A total of 131 lncRNAs, 185 miRNA, and 1309 mRNAs were found to be differentially expressed in AFP-negative HCC. A ceRNA network consisting of 12 lncRNA, 23 miRNA, and 74 mRNA was constructed. The top ten hub genes including EZH2, CCNB1, E2F1, PBK, CHAF1A, ESR1, RRM2, CCNE1, MCM4, and ATAD2 showed good diagnostic power under the ROC curve; and 2 lncRNAs (LINC00261, LINC00482), 3 miRNAs (hsa-miR-93, hsa-miR-221, hsa-miR-222), and 2 mRNAs (EGR2, LPCAT1) were found to be associated with the overall survival of AFP-negative patients.
Conclusion: This study could provide a novel insight into the molecular pathogenesis of AFP-negative HCC and reveal some candidate diagnostic and prognostic biomarkers for AFP-negative HCC.
Keywords: AFP-negative HCC; ceRNA network; diagnosis; prognosis.
© 2021 Liu et al.
Conflict of interest statement
The authors declared no conflicts of interest for this work and no competing financial interests.
Figures







References
-
- Cui DJ, Wu Y, Wen DH. CD34, PCNA and CK19 expressions in AFP- hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(16):5200–5205. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

