Gut microbiome is associated with multiple sclerosis activity in children
- PMID: 34409759
- PMCID: PMC8419410
- DOI: 10.1002/acn3.51441
Gut microbiome is associated with multiple sclerosis activity in children
Abstract
Objective: To identify features of the gut microbiome associated with multiple sclerosis activity over time.
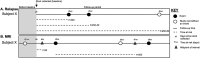
Methods: We used 16S ribosomal RNA sequencing from stool of 55 recently diagnosed pediatric-onset multiple sclerosis patients. Microbiome features included the abundance of individual microbes and networks identified from weighted genetic correlation network analyses. Prentice-Williams-Peterson Cox proportional hazards models estimated the associations between features and three disease activity outcomes: clinical relapses and both new/enlarging T2 lesions and new gadolinium-enhancing lesions on brain MRI. Analyses were adjusted for age, sex, and disease-modifying therapies.
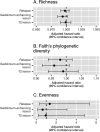
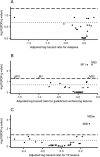
Results: Participants were followed, on average, 2.1 years. Five microbes were nominally associated with all three disease activity outcomes after multiple testing correction. These included butyrate producers Odoribacter (relapse hazard ratio = 0.46, 95% confidence interval: 0.24, 0.88) and Butyricicoccus (relapse hazard ratio = 0.49, 95% confidence interval: 0.28, 0.88). Two networks of co-occurring gut microbes were significantly associated with a higher hazard of both MRI outcomes (gadolinium-enhancing lesion hazard ratios (95% confidence intervals) for Modules 32 and 33 were 1.29 (1.08, 1.54) and 1.42 (1.18, 1.71), respectively; T2 lesion hazard ratios (95% confidence intervals) for Modules 32 and 33 were 1.34 (1.15, 1.56) and 1.41 (1.21, 1.64), respectively). Metagenomic predictions of these networks demonstrated enrichment for amino acid biosynthesis pathways.
Interpretation: Both individual and networks of gut microbes were associated with longitudinal multiple sclerosis activity. Known functions and metagenomic predictions of these microbes suggest the important role of butyrate and amino acid biosynthesis pathways. This provides strong support for future development of personalized microbiome interventions to modify multiple sclerosis disease activity.
© 2021 The Authors. Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
The authors report no competing interests.
Figures



References
-
- Dopkins N, Nagarkatti PS, Nagarkatti M. The role of gut microbiome and associated metabolome in the regulation of neuroinflammation in multiple sclerosis and its implications in attenuating chronic inflammation in other inflammatory and autoimmune disorders. Immunology 2018;154(2):178–185. 10.1111/imm.12903 - DOI - PMC - PubMed

