Longitudinal K-means approaches to clustering and analyzing EHR opioid use trajectories for clinical subtypes
- PMID: 34411708
- PMCID: PMC9035269
- DOI: 10.1016/j.jbi.2021.103889
Longitudinal K-means approaches to clustering and analyzing EHR opioid use trajectories for clinical subtypes
Abstract
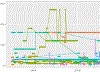
Identification of patient subtypes from retrospective Electronic Health Record (EHR) data is fraught with inherent modeling issues, such as missing data and variable length time intervals, and the results obtained are highly dependent on data pre-processing strategies. As we move towards personalized medicine, assessing accurate patient subtypes will be a key factor in creating patient specific treatment plans. Partitioning longitudinal trajectories from irregularly spaced and variable length time intervals is a well-established, but open problem. In this work, we present and compare k-means approaches for subtyping opioid use trajectories from EHR data. We then interpret the resulting subtypes using decision trees, examining how each subtype is influenced by opioid medication features and patient diagnoses, procedures, and demographics. Finally, we discuss how the subtypes can be incorporated in static machine learning models as features in predicting opioid overdose and adverse events. The proposed methods are general, and can be extended to other EHR prescription dosage trajectories.
Keywords: Electronic health records; Longitudinal k-means clustering; Opioids; Patient subtypes; Trajectory analysis.
Copyright © 2021. Published by Elsevier Inc.
Figures



References
-
- Van Calster B, Wynants L. Machine Learning in Medicine. New England Journal Of Medicine. 2019;380(26):2588-. - PubMed
-
- Aghabozorgi S, Shirkhorshidi AS, Wah TY. Time-series clustering–a decade review. Information Systems. 2015;53:16–38.
-
- Schulam P, Arora R, editors. Disease trajectory maps. Advances in neural information processing systems; 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

