Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: Results and implications from a collaborative inter-laboratory study in Canada
- PMID: 34412784
- PMCID: PMC7929783
- DOI: 10.1016/j.jes.2021.01.029
Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: Results and implications from a collaborative inter-laboratory study in Canada
Abstract
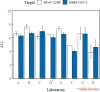
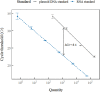
Detection of SARS-CoV-2 RNA in wastewater is a promising tool for informing public health decisions during the COVID-19 pandemic. However, approaches for its analysis by use of reverse transcription quantitative polymerase chain reaction (RT-qPCR) are still far from standardized globally. To characterize inter- and intra-laboratory variability among results when using various methods deployed across Canada, aliquots from a real wastewater sample were spiked with surrogates of SARS-CoV-2 (gamma-radiation inactivated SARS-CoV-2 and human coronavirus strain 229E [HCoV-229E]) at low and high levels then provided "blind" to eight laboratories. Concentration estimates reported by individual laboratories were consistently within a 1.0-log10 range for aliquots of the same spiked condition. All laboratories distinguished between low- and high-spikes for both surrogates. As expected, greater variability was observed in the results amongst laboratories than within individual laboratories, but SARS-CoV-2 RNA concentration estimates for each spiked condition remained mostly within 1.0-log10 ranges. The no-spike wastewater aliquots provided yielded non-detects or trace levels (<20 gene copies/mL) of SARS-CoV-2 RNA. Detections appear linked to methods that included or focused on the solids fraction of the wastewater matrix and might represent in-situ SARS-CoV-2 to the wastewater sample. HCoV-229E RNA was not detected in the no-spike aliquots. Overall, all methods yielded comparable results at the conditions tested. Partitioning behavior of SARS-CoV-2 and spiked surrogates in wastewater should be considered to evaluate method effectiveness. A consistent method and laboratory to explore wastewater SARS-CoV-2 temporal trends for a given system, with appropriate quality control protocols and documented in adequate detail should succeed.
Keywords: COVID-19; Public health; Quality assurance; Quality control; Wastewater surveillance.
Copyright © 2021. Published by Elsevier B.V.
Figures





References
-
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. - DOI - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. - DOI - PMC - PubMed
-
- Alpaslan Kocamemi B., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. MedRxiv. 2020 doi: 10.1101/2020.05.12.20099358. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

