Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1
- PMID: 34414377
- PMCID: PMC8373048
- DOI: 10.1038/s43018-020-0071-1
Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1
Erratum in
-
Publisher Correction: Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1.Nat Cancer. 2020 Sep;1(9):935. doi: 10.1038/s43018-020-00118-z. Nat Cancer. 2020. PMID: 35121957 No abstract available.
Abstract
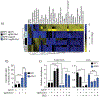
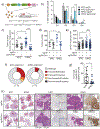
Approximately 20-30% of human lung adenocarcinomas (LUAD) harbor loss-of-function (LOF) mutations in Kelch-like ECH Associated-Protein 1 (KEAP1), which lead to hyperactivation of the nuclear factor, erythroid 2-like 2 (NRF2) antioxidant pathway and correlate with poor prognosis1-3. We previously showed that Keap1 mutation accelerates KRAS-driven LUAD and produces a marked dependency on glutaminolysis4. To extend the investigation of genetic dependencies in the context of Keap1 mutation, we performed a druggable genome CRISPR-Cas9 screen in Keap1-mutant cells. This analysis uncovered a profound Keap1 mutant-specific dependency on solute carrier family 33 member 1 (Slc33a1), an endomembrane-associated protein with roles in autophagy regulation5, as well as a series of functionally-related genes implicated in the unfolded protein response. Targeted genetic and biochemical experiments using mouse and human Keap1-mutant tumor lines, as well as preclinical genetically-engineered mouse models (GEMMs) of LUAD, validate Slc33a1 as a robust Keap1-mutant-specific dependency. Furthermore, unbiased genome-wide CRISPR screening identified additional genes related to Slc33a1 dependency. Overall, our study provides a strong rationale for stratification of patients harboring KEAP1-mutant or NRF2-hyperactivated tumors as likely responders to targeted SLC33A1 inhibition and underscores the value of integrating functional genetic approaches with GEMMs to identify and validate genotype-specific therapeutic targets.
Conflict of interest statement
Declaration of Interest: T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific. He is also a co-Founder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. T.J. laboratory currently also receives funding from the Johnson & Johnson Lung Cancer Initiative, but this funding did not support the research described in this manuscript. This work was supported by the Howard Hughes Medical Institute and Calico Life Sciences LLC.
Figures





Comment in
-
Keap1ing an eye on Slc33A1.Nat Cancer. 2020 Jun;1(6):575-576. doi: 10.1038/s43018-020-0072-0. Nat Cancer. 2020. PMID: 35121976 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

