A monocyte/dendritic cell molecular signature of SARS-CoV-2-related multisystem inflammatory syndrome in children with severe myocarditis
- PMID: 34414385
- PMCID: PMC8363470
- DOI: 10.1016/j.medj.2021.08.002
A monocyte/dendritic cell molecular signature of SARS-CoV-2-related multisystem inflammatory syndrome in children with severe myocarditis
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children is generally milder than in adults, but a proportion of cases result in hyperinflammatory conditions often including myocarditis.
Methods: To better understand these cases, we applied a multiparametric approach to the study of blood cells of 56 children hospitalized with suspicion of SARS-CoV-2 infection. Plasma cytokine and chemokine levels and blood cellular composition were measured, alongside gene expression at the bulk and single-cell levels.
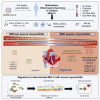
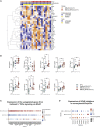
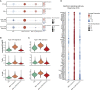
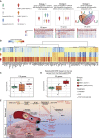
Findings: The most severe forms of multisystem inflammatory syndrome in children (MIS-C) related to SARS-CoV-2 that resulted in myocarditis were characterized by elevated levels of pro-angiogenesis cytokines and several chemokines. Single-cell transcriptomics analyses identified a unique monocyte/dendritic cell gene signature that correlated with the occurrence of severe myocarditis characterized by sustained nuclear factor κB (NF-κB) activity and tumor necrosis factor alpha (TNF-α) signaling and associated with decreased gene expression of NF-κB inhibitors. We also found a weak response to type I and type II interferons, hyperinflammation, and response to oxidative stress related to increased HIF-1α and Vascular endothelial growth factor (VEGF) signaling.
Conclusions: These results provide potential for a better understanding of disease pathophysiology.
Funding: Agence National de la Recherche (Institut Hospitalo-Universitaire Imagine, grant ANR-10-IAHU-01; Recherche Hospitalo-Universitaire, grant ANR-18-RHUS-0010; Laboratoire d'Excellence ''Milieu Intérieur," grant ANR-10-LABX-69-01; ANR-flash Covid19 "AIROCovid" and "CoVarImm"), Institut National de la Santé et de la Recherche Médicale (INSERM), and the "URGENCE COVID-19" fundraising campaign of Institut Pasteur.
Keywords: COVID-19; Kawasaki Disease; MIS-C; PIMS-TS; SARS-CoV-2; TNF-α and NF-κB signaling; lack of responses to type I and type II IFN secretion; myocarditis; scRNA-seq.
© 2021 Elsevier Inc.
Conflict of interest statement
D.D., F.R.-L., J.T., and M.M.M. are listed as inventors on a patent application related to this technology (European Patent Application no. EP21305197, entitled “Methods of predicting multisystem inflammatory syndrome [MIS-C] with severe myocarditis in subjects suffering from a SARS-CoV-2 infection”).
Figures




Comment in
-
The Heart of the Problem in Multisystem Inflammatory Syndrome in Children.Med. 2021 Sep 10;2(9):993-995. doi: 10.1016/j.medj.2021.08.008. Med. 2021. PMID: 34522906 Free PMC article.
References
-
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782–793. - PubMed
-
- Brodin P. Why is COVID-19 so mild in children? Acta Paediatr. 2020;109:1082–1083. - PubMed
-
- Castagnoli R., Votto M., Licari A., Brambilla I., Bruno R., Perlini S., Rovida F., Baldanti F., Marseglia G.L. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection in Children and Adolescents: A Systematic Review. JAMA Pediatr. 2020;174:882–889. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

