Reviewing and assessing existing meta-analysis models and tools
- PMID: 34415019
- PMCID: PMC8575034
- DOI: 10.1093/bib/bbab324
Reviewing and assessing existing meta-analysis models and tools
Abstract
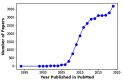
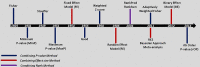
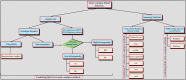
Over the past few years, meta-analysis has become popular among biomedical researchers for detecting biomarkers across multiple cohort studies with increased predictive power. Combining datasets from different sources increases sample size, thus overcoming the issue related to limited sample size from each individual study and boosting the predictive power. This leads to an increased likelihood of more accurately predicting differentially expressed genes/proteins or significant biomarkers underlying the biological condition of interest. Currently, several meta-analysis methods and tools exist, each having its own strengths and limitations. In this paper, we survey existing meta-analysis methods, and assess the performance of different methods based on results from different datasets as well as assessment from prior knowledge of each method. This provides a reference summary of meta-analysis models and tools, which helps to guide end-users on the choice of appropriate models or tools for given types of datasets and enables developers to consider current advances when planning the development of new meta-analysis models and more practical integrative tools.
Keywords: cohort study; data integration; experimental study; meta-analysis; predictive power; sample size.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Mazandu GK, Kyomugisha I, Geza E, et al. Designing data-driven learning algorithms: A necessity to ensure effective post-genomic medicine and biomedical research. In: Artifi-cial Intelligence - Applications in Medicine and Biology. 5 Princes Gate Court. London, UK: IntechOpen Publisher, 2019, 3–18.
-
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8(1):118–27. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

