tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes
- PMID: 34417604
- PMCID: PMC8450103
- DOI: 10.1093/nar/gkab688
tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes
Abstract
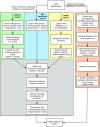
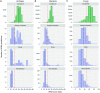
tRNAscan-SE has been widely used for transfer RNA (tRNA) gene prediction for over twenty years, developed just as the first genomes were decoded. With the massive increase in quantity and phylogenetic diversity of genomes, the accurate detection and functional prediction of tRNAs has become more challenging. Utilizing a vastly larger training set, we created nearly one hundred specialized isotype- and clade-specific models, greatly improving tRNAscan-SE's ability to identify and classify both typical and atypical tRNAs. We employ a new comparative multi-model strategy where predicted tRNAs are scored against a full set of isotype-specific covariance models, allowing functional prediction based on both the anticodon and the highest-scoring isotype model. Comparative model scoring has also enhanced the program's ability to detect tRNA-derived SINEs and other likely pseudogenes. For the first time, tRNAscan-SE also includes fast and highly accurate detection of mitochondrial tRNAs using newly developed models. Overall, tRNA detection sensitivity and specificity is improved for all isotypes, particularly those utilizing specialized models for selenocysteine and the three subtypes of tRNA genes encoding a CAU anticodon. These enhancements will provide researchers with more accurate and detailed tRNA annotation for a wider variety of tRNAs, and may direct attention to tRNAs with novel traits.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

