ENDO-Pore: high-throughput linked-end mapping of single DNA cleavage events using nanopore sequencing
- PMID: 34417616
- PMCID: PMC8599736
- DOI: 10.1093/nar/gkab727
ENDO-Pore: high-throughput linked-end mapping of single DNA cleavage events using nanopore sequencing
Abstract
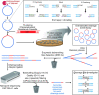
Mapping the precise position of DNA cleavage events plays a key role in determining the mechanism and function of endonucleases. ENDO-Pore is a high-throughput nanopore-based method that allows the time resolved mapping single molecule DNA cleavage events in vitro. Following linearisation of a circular DNA substrate by the endonuclease, a resistance cassette is ligated recording the position of the cleavage event. A library of single cleavage events is constructed and subjected to rolling circle amplification to generate concatemers. These are sequenced and used to produce accurate consensus sequences. To identify the cleavage site(s), we developed CSI (Cleavage Site Investigator). CSI recognizes the ends of the cassette ligated into the cleaved substrate and triangulates the position of the dsDNA break. We firstly benchmarked ENDO-Pore using Type II restriction endonucleases. Secondly, we analysed the effect of crRNA length on the cleavage pattern of CRISPR Cas12a. Finally, we mapped the time-resolved DNA cleavage by the Type ISP restriction endonuclease LlaGI that introduces random double-strand breaks into its DNA substrates.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

