HIV-1 Tat and cocaine impact astrocytic energy reservoir influence on miRNA epigenetic regulation
- PMID: 34418497
- PMCID: PMC8671230
- DOI: 10.1016/j.ygeno.2021.08.013
HIV-1 Tat and cocaine impact astrocytic energy reservoir influence on miRNA epigenetic regulation
Abstract
Astrocytes are the primary regulator of energy metabolism in the central nervous system (CNS), and impairment of astrocyte's energy resource may trigger neurodegeneration. HIV infections and cocaine use are known to alter epigenetic modification, including miRNAs, which can target gene expression post-transcriptionally. However, miRNA-mediated astrocyte energy metabolism has not been delineated in HIV infection and cocaine abuse. Using next-generation sequencing (NGS), we identified a total of 1900 miRNAs, 64 were upregulated and 68 miRNAs were downregulated in the astrocytes by HIV-1 Tat with cocaine exposure. Moreover, miR-4727-3p, miR-5189-5p, miR-5090, and miR-6810-5p expressions were significantly impacted, and their gene targets were identified as VAMP2, NFIB, PPM1H, MEIS1, and PSD93 through the bioinformatic approach. In addition, the astrocytes treated with the nootropic drug piracetam protects these miRNAs. These findings provide evidence that the miRNAs in the astrocytes may be a potential biomarker and therapeutic target for HIV and cocaine abuse-induced neurodegeneration.
Keywords: Astrocyte and cocaine; HIV-1 Tat; Piracetam; miRNA.
Copyright © 2021 Elsevier Inc. All rights reserved.
Figures





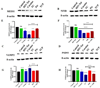
References
-
- World Health Organization, U.S. Statistics | HIV.gov, Underst. Addressing Violence against Women. (2018) 1–12. https://www.hiv.gov/hiv-basics/overview/data-and-trends/statistics (accessed January 13, 2021).
-
- Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, Toossi Z, Colebunders RL, Jensen MA, Mullins JI, Vanham G, Arts EJ, Changes in Human Immunodeficiency Virus Type 1 Fitness and Genetic Diversity during Disease Progression, J. Virol 79 (2005) 9006–9018. 10.1128/jvi.79.14.9006-9018.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

