Mucosa-Associated Microbial Profile Is Altered in Small Intestinal Bacterial Overgrowth
- PMID: 34421869
- PMCID: PMC8372370
- DOI: 10.3389/fmicb.2021.710940
Mucosa-Associated Microbial Profile Is Altered in Small Intestinal Bacterial Overgrowth
Abstract
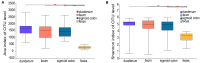
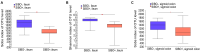
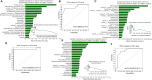
The overall gut microbial profile of patients with small intestinal bacterial overgrowth (SIBO) has not been thoroughly investigated. We investigated the microbial communities of mucosal specimens from the duodenum, ileum, sigmoid colon, and feces of patients with and without SIBO, as diagnosed by lactulose breath testing. The bacteria present in the mucosal and fecal samples were identified using 16S rRNA gene sequencing. Further analysis was performed using the linear discriminant analysis (LDA) effect size method, random forest analysis, and receiver operating characteristic analysis. The microbial diversities of the fecal samples were significantly lower than those of the mucosal samples from the duodenum, ileum, and sigmoid colon (P < 0.001, P < 0.001, and P < 0.001, respectively), while the bacterial compositions of the ileac mucosal samples and sigmoid mucosal samples were similar. The bacterial composition of either the fecal or duodenal mucosal samples were significantly different from those of the other three groups (ANOSIM R = 0.305, P = 0.001). The bacterial compositions of the mucosal samples of the duodenum, ileum, and sigmoid colon in the SIBO + subjects were significantly different from those of the SIBO- subjects (ANOSIM P = 0.039, 0.002, and 0.007, respectively). The relative abundances of 7, 18, and 8 genera were significantly different (LDA score > 3) in the mucosal samples of the duodenum, ileum, and sigmoid colon between the SIBO + and SIBO- groups. Four genera (Lactobacillus, Prevotella_1, Dialister, and norank_f__Ruminococcaceae) showed similar changes among the mucosal samples of the duodenum, ileum, and sigmoid colon in the SIBO + subjects. A signature consisting of four genera in the duodenal mucosa, three genera in the ileac mucosa, or six genera in the mucosa of the sigmoid colon exhibited predictive power for SIBO (area under the curve = 0.9, 0.93, and 0.87, respectively). This study provides a comprehensive profile of the gut microbiota in patients with SIBO. Dysbiosis was observed in the mucosa-associated gut microbiome but not in the fecal microbiome of patients with SIBO. Furthermore, we identified mucosa-associated taxa that may be potential biomarkers or therapeutic targets of SIBO. Further investigation is needed on their mechanisms and roles in SIBO.
Keywords: 16S rRNA gene sequencing; SIBO; biomarker; duodenum; ileum; microbiota; mucosa.
Copyright © 2021 Li, Zhang, Ma, Tang, Li, Li and Wan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Carroll I. M., Ringel-Kulka T., Keku T. O., Chang Y. H., Packey C. D., Sartor R. B., et al. (2011). Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 301 G799–G807. 10.1152/ajpgi.00154.2011 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

