HIV-2 diversity displays two clades within group A with distinct geographical distribution and evolution
- PMID: 34422316
- PMCID: PMC8377049
- DOI: 10.1093/ve/veab024
HIV-2 diversity displays two clades within group A with distinct geographical distribution and evolution
Abstract
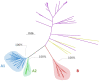
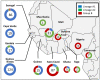
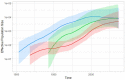
Genetic diversity of HIV-2 groups A and B has not yet been fully described, especially in a few Western Africa countries such as Ivory-Coast or Mali. We collected 444 pol, 152 vif, 129 env, and 74 LTR sequences from patients of the French ANRS CO5 HIV-2 cohort completed by 221 pol, 18 vif, 377 env, and 63 LTR unique sequences from public databases. We performed phylogenetic reconstructions and revealed two distinct lineages within HIV-2 group A, herein called A1 and A2, presenting non-negligible genetic distances and distinct geographic distributions as A1 is related to coastal Western African countries and A2 to inland Western countries. Estimated early diversification times for groups A and B in human populations were 1940 [95% higher probability densitiy: 1935-53] and 1961 [1952-70]. A1 experienced an early diversification in 1942 [1937-58] with two distinct early epidemics in Guinea-Bissau or Senegal, raising the possibility of group A emergence in those countries from an initial introduction from Ivory-Coast to Senegal, two former French colonies. Changes in effective population sizes over time revealed that A1 exponentially grew concomitantly to Guinea-Bissau independence war, but both A2 and B lineages experienced a latter growth, starting during the 80s economic crisis. This large HIV-2 genetic analysis provides the existence of two distinct subtypes within group A and new data about HIV-2 early spreading patterns and recent epidemiologic evolution for which data are scarce outside Guinea-Bissau.
Keywords: HIV-2; molecular epidemiology; phylogenetic; viral diversity.
© The Author(s) 2021. Published by Oxford University Press.
Figures


References
LinkOut - more resources
Full Text Sources

