When Viruses Cross Developmental Pathways
- PMID: 34422814
- PMCID: PMC8375270
- DOI: 10.3389/fcell.2021.691644
When Viruses Cross Developmental Pathways
Abstract
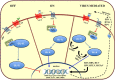
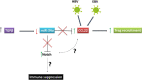
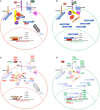
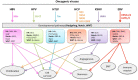
Aberrant regulation of developmental pathways plays a key role in tumorigenesis. Tumor cells differ from normal cells in their sustained proliferation, replicative immortality, resistance to cell death and growth inhibition, angiogenesis, and metastatic behavior. Often they acquire these features as a consequence of dysregulated Hedgehog, Notch, or WNT signaling pathways. Human tumor viruses affect the cancer cell hallmarks by encoding oncogenic proteins, and/or by modifying the microenvironment, as well as by conveying genomic instability to accelerate cancer development. In addition, viral immune evasion mechanisms may compromise developmental pathways to accelerate tumor growth. Viruses achieve this by influencing both coding and non-coding gene regulatory pathways. Elucidating how oncogenic viruses intersect with and modulate developmental pathways is crucial to understanding viral tumorigenesis. Many currently available antiviral therapies target viral lytic cycle replication but with low efficacy and severe side effects. A greater understanding of the cross-signaling between oncogenic viruses and developmental pathways will improve the efficacy of next-generation inhibitors and pave the way to more targeted antiviral therapies.
Keywords: Hedgehog; Notch; WNT; immune evasion; microRNA; oncogenic viruses; targeted therapies.
Copyright © 2021 Trivedi, Patel, Bellavia, Messina, Palermo, Ceccarelli, Marchese, Anastasiadou, Minter and Felli.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

