DNA-Encoded Chemical Libraries: A Comprehensive Review with Succesful Stories and Future Challenges
- PMID: 34423264
- PMCID: PMC8369695
- DOI: 10.1021/acsptsci.1c00118
DNA-Encoded Chemical Libraries: A Comprehensive Review with Succesful Stories and Future Challenges
Abstract
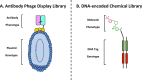
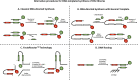
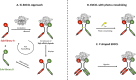
DNA-encoded chemical libraries (DELs) represent a versatile and powerful technology platform for the discovery of small-molecule ligands to protein targets of biological and pharmaceutical interest. DELs are collections of molecules, individually coupled to distinctive DNA tags serving as amplifiable identification barcodes. Thanks to advances in DNA-compatible reactions, selection methodologies, next-generation sequencing, and data analysis, DEL technology allows the construction and screening of libraries of unprecedented size, which has led to the discovery of highly potent ligands, some of which have progressed to clinical trials. In this Review, we present an overview of diverse approaches for the generation and screening of DEL molecular repertoires. Recent success stories are described, detailing how novel ligands were isolated from DEL screening campaigns and were further optimized by medicinal chemistry. The goal of the Review is to capture some of the most recent developments in the field, while also elaborating on future challenges to further improve DEL technology as a therapeutic discovery platform.
© 2021 American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): D.N. is a cofounder and shareholder of Philogen (www.philogen.com), a Swiss-Italian biotech company that operates in the field of ligand-based pharmacodelivery. A.G.-M., E.J.D., and F.S. are employees of Philochem AG, the daughter company of Philogen, acting as discovery unit of the group.
Figures




References
-
- Macarron R.; Banks M. N.; Bojanic D.; Burns D. J.; Cirovic D. A.; Garyantes T.; Green D. V. S.; Hertzberg R. P.; Janzen W. P.; Paslay J. W.; Schopfer U.; Sittampalam G. S. (2011) Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discovery 10, 188–195. 10.1038/nrd3368. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
