Insertion of atypical glycans into the tumor antigen-binding site identifies DLBCLs with distinct origin and behavior
- PMID: 34424958
- PMCID: PMC8554650
- DOI: 10.1182/blood.2021012052
Insertion of atypical glycans into the tumor antigen-binding site identifies DLBCLs with distinct origin and behavior
Abstract
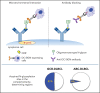
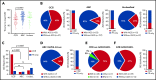
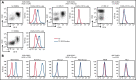
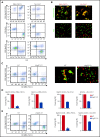
Glycosylation of the surface immunoglobulin (Ig) variable region is a remarkable follicular lymphoma-associated feature rarely seen in normal B cells. Here, we define a subset of diffuse large B-cell lymphomas (DLBCLs) that acquire N-glycosylation sites selectively in the Ig complementarity-determining regions (CDRs) of the antigen-binding sites. Mass spectrometry and X-ray crystallography demonstrate how the inserted glycans are stalled at oligomannose-type structures because they are buried in the CDR loops. Acquisition of sites occurs in ∼50% of germinal-center B-cell-like DLBCL (GCB-DLBCL), mainly of the genetic EZB subtype, irrespective of IGHV-D-J use. This markedly contrasts with the activated B-cell-like DLBCL Ig, which rarely has sites in the CDR and does not seem to acquire oligomannose-type structures. Acquisition of CDR-located acceptor sites associates with mutations of epigenetic regulators and BCL2 translocations, indicating an origin shared with follicular lymphoma. Within the EZB subtype, these sites are associated with more rapid disease progression and with significant gene set enrichment of the B-cell receptor, PI3K/AKT/MTORC1 pathway, glucose metabolism, and MYC signaling pathways, particularly in the fraction devoid of MYC translocations. The oligomannose-type glycans on the lymphoma cells interact with the candidate lectin dendritic cell-specific intercellular adhesion molecule 3 grabbing non-integrin (DC-SIGN), mediating low-level signals, and lectin-expressing cells form clusters with lymphoma cells. Both clustering and signaling are inhibited by antibodies specifically targeting the DC-SIGN carbohydrate recognition domain. Oligomannosylation of the tumor Ig is a posttranslational modification that readily identifies a distinct GCB-DLBCL category with more aggressive clinical behavior, and it could be a potential precise therapeutic target via antibody-mediated inhibition of the tumor Ig interaction with DC-SIGN-expressing M2-polarized macrophages.
© 2021 by The American Society of Hematology.
Figures



Comment in
-
Sugar-modified B-cell receptors in DLBCL.Blood. 2021 Oct 28;138(17):1512-1514. doi: 10.1182/blood.2021013629. Blood. 2021. PMID: 34709382 No abstract available.
References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. . Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769): 503-511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, et al. ; Lymphoma/Leukemia Molecular Profiling Project . The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937-1947. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

