The genetic structure of the Turkish population reveals high levels of variation and admixture
- PMID: 34426522
- PMCID: PMC8433500
- DOI: 10.1073/pnas.2026076118
The genetic structure of the Turkish population reveals high levels of variation and admixture
Erratum in
-
Correction for Kars et al., The genetic structure of the Turkish population reveals high levels of variation and admixture.Proc Natl Acad Sci U S A. 2021 Dec 28;118(52):e2120031118. doi: 10.1073/pnas.2120031118. Proc Natl Acad Sci U S A. 2021. PMID: 34949644 Free PMC article. No abstract available.
Abstract
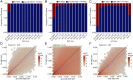
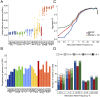
The construction of population-based variomes has contributed substantially to our understanding of the genetic basis of human inherited disease. Here, we investigated the genetic structure of Turkey from 3,362 unrelated subjects whose whole exomes (n = 2,589) or whole genomes (n = 773) were sequenced to generate a Turkish (TR) Variome that should serve to facilitate disease gene discovery in Turkey. Consistent with the history of present-day Turkey as a crossroads between Europe and Asia, we found extensive admixture between Balkan, Caucasus, Middle Eastern, and European populations with a closer genetic relationship of the TR population to Europeans than hitherto appreciated. We determined that 50% of TR individuals had high inbreeding coefficients (≥0.0156) with runs of homozygosity longer than 4 Mb being found exclusively in the TR population when compared to 1000 Genomes Project populations. We also found that 28% of exome and 49% of genome variants in the very rare range (allele frequency < 0.005) are unique to the modern TR population. We annotated these variants based on their functional consequences to establish a TR Variome containing alleles of potential medical relevance, a repository of homozygous loss-of-function variants and a TR reference panel for genotype imputation using high-quality haplotypes, to facilitate genome-wide association studies. In addition to providing information on the genetic structure of the modern TR population, these data provide an invaluable resource for future studies to identify variants that are associated with specific phenotypes as well as establishing the phenotypic consequences of mutations in specific genes.
Keywords: Turkish Variome; admixture; population genetics; sequencing; variation.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Skourtanioti E., et al. ., Genomic history of neolithic to bronze age Anatolia, northern Levant, and southern Caucasus. Cell 181, 1158–1175.e28 (2020). - PubMed
-
- Golden P. B., An Introduction to the History of the Turkic Peoples. Ethnogenesis and State-Formation in Medieval and Early Modern Eurasia and the Middle East (Otto Harrassowitz, Wiesbaden, 1992).
-
- Cinnioğlu C., et al. ., Excavating Y-chromosome haplotype strata in Anatolia. Hum. Genet. 114, 127–148 (2004). - PubMed
-
- Akbayram S., et al. ., The frequency of consanguineous marriage in eastern Turkey. Genet. Couns. 20, 207–214 (2009). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

