Ankyrin repeats in context with human population variation
- PMID: 34428215
- PMCID: PMC8415598
- DOI: 10.1371/journal.pcbi.1009335
Ankyrin repeats in context with human population variation
Abstract
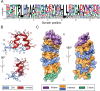
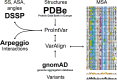
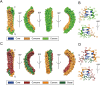
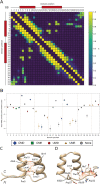
Ankyrin protein repeats bind to a wide range of substrates and are one of the most common protein motifs in nature. Here, we collate a high-quality alignment of 7,407 ankyrin repeats and examine for the first time, the distribution of human population variants from large-scale sequencing of healthy individuals across this family. Population variants are not randomly distributed across the genome but are constrained by gene essentiality and function. Accordingly, we interpret the population variants in context with evolutionary constraint and structural features including secondary structure, accessibility and protein-protein interactions across 383 three-dimensional structures of ankyrin repeats. We find five positions that are highly conserved across homologues and also depleted in missense variants within the human population. These positions are significantly enriched in intra-domain contacts and so likely to be key for repeat packing. In contrast, a group of evolutionarily divergent positions are found to be depleted in missense variants in human and significantly enriched in protein-protein interactions. Our analysis also suggests the domain has three, not two surfaces, each with different patterns of enrichment in protein-substrate interactions and missense variants. Our findings will be of interest to those studying or engineering ankyrin-repeat containing proteins as well as those interpreting the significance of disease variants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- BB/J01446X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 102132/B/13/Z/WT_/Wellcome Trust/United Kingdom
- BB/J019364/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G022682/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

