Evaluating the performance of a clinical genome sequencing program for diagnosis of rare genetic disease, seen through the lens of craniosynostosis
- PMID: 34429528
- PMCID: PMC8629760
- DOI: 10.1038/s41436-021-01297-5
Evaluating the performance of a clinical genome sequencing program for diagnosis of rare genetic disease, seen through the lens of craniosynostosis
Abstract
Purpose: Genome sequencing (GS) for diagnosis of rare genetic disease is being introduced into the clinic, but the complexity of the data poses challenges for developing pipelines with high diagnostic sensitivity. We evaluated the performance of the Genomics England 100,000 Genomes Project (100kGP) panel-based pipelines, using craniosynostosis as a test disease.
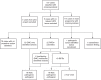
Methods: GS data from 114 probands with craniosynostosis and their relatives (314 samples), negative on routine genetic testing, were scrutinized by a specialized research team, and diagnoses compared with those made by 100kGP.
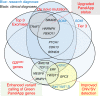
Results: Sixteen likely pathogenic/pathogenic variants were identified by 100kGP. Eighteen additional likely pathogenic/pathogenic variants were identified by the research team, indicating that for craniosynostosis, 100kGP panels had a diagnostic sensitivity of only 47%. Measures that could have augmented diagnoses were improved calling of existing panel genes (+18% sensitivity), review of updated panels (+12%), comprehensive analysis of de novo small variants (+29%), and copy-number/structural variants (+9%). Recent NHS England recommendations that partially incorporate these measures should achieve 85% overall sensitivity (+38%).
Conclusion: GS identified likely pathogenic/pathogenic variants in 29.8% of previously undiagnosed patients with craniosynostosis. This demonstrates the value of research analysis and the importance of continually improving algorithms to maximize the potential of clinical GS.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Genomics England. The 100,000 Genomes Project protocol. 2017. https://figshare.com/articles/journal_contribution/GenomicEnglandProtoco.... Accessed 14 June 2021.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

