The roles of history, chance, and natural selection in the evolution of antibiotic resistance
- PMID: 34431477
- PMCID: PMC8412936
- DOI: 10.7554/eLife.70676
The roles of history, chance, and natural selection in the evolution of antibiotic resistance
Abstract
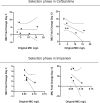
History, chance, and selection are the fundamental factors that drive and constrain evolution. We designed evolution experiments to disentangle and quantify effects of these forces on the evolution of antibiotic resistance. Previously, we showed that selection of the pathogen Acinetobacter baumannii in both structured and unstructured environments containing the antibiotic ciprofloxacin produced distinct genotypes and phenotypes, with lower resistance in biofilms as well as collateral sensitivity to β-lactam drugs (Santos-Lopez et al., 2019). Here we study how this prior history influences subsequent evolution in new β-lactam antibiotics. Selection was imposed by increasing concentrations of ceftazidime and imipenem and chance differences arose as random mutations among replicate populations. The effects of history were reduced by increasingly strong selection in new drugs, but not erased, at times revealing important contingencies. A history of selection in structured environments constrained resistance to new drugs and led to frequent loss of resistance to the initial drug by genetic reversions and not compensatory mutations. This research demonstrates that despite strong selective pressures of antibiotics leading to genetic parallelism, history can etch potential vulnerabilities to orthogonal drugs.
Keywords: acinetobacter; collateral sensitivity; efflux; evolutionary biology; genomics; infectious disease; microbiology; none; population genetics.
© 2021, Santos-Lopez et al.
Conflict of interest statement
AS, CM, CT, JR, VC None, AH none
Figures







References
-
- Alonso-del Valle A, León-Sampedro R, Rodríguez-Beltrán J, DelaFuente J, Hernández-García M, Ruiz-Garbajosa P, Cantón R, Peña-Miller R, San Millán A. Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities. Nature Communications. 2021;12:2653. doi: 10.1038/s41467-021-22849-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

