Mechanistic Insights into the Capsule-Targeting Depolymerase from a Klebsiella pneumoniae Bacteriophage
- PMID: 34431721
- PMCID: PMC8552709
- DOI: 10.1128/Spectrum.01023-21
Mechanistic Insights into the Capsule-Targeting Depolymerase from a Klebsiella pneumoniae Bacteriophage
Abstract
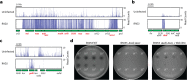
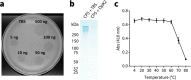
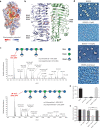
The production of capsular polysaccharides by Klebsiella pneumoniae protects the bacterial cell from harmful environmental factors such as antimicrobial compounds and infection by bacteriophages (phages). To bypass this protective barrier, some phages encode polysaccharide-degrading enzymes referred to as depolymerases to provide access to cell surface receptors. Here, we characterized the phage RAD2, which infects K. pneumoniae strains that produce the widespread, hypervirulence-associated K2-type capsular polysaccharide. Using transposon-directed insertion sequencing, we have shown that the production of capsule is an absolute requirement for efficient RAD2 infection by serving as a first-stage receptor. We have identified the depolymerase responsible for recognition and degradation of the capsule, determined that the depolymerase forms globular appendages on the phage virion tail tip, and present the cryo-electron microscopy structure of the RAD2 capsule depolymerase at 2.7-Å resolution. A putative active site for the enzyme was identified, comprising clustered negatively charged residues that could facilitate the hydrolysis of target polysaccharides. Enzymatic assays coupled with mass spectrometric analyses of digested oligosaccharide products provided further mechanistic insight into the hydrolase activity of the enzyme, which, when incubated with K. pneumoniae, removes the capsule and sensitizes the cells to serum-induced killing. Overall, these findings expand our understanding of how phages target the Klebsiella capsule for infection, providing a framework for the use of depolymerases as antivirulence agents against this medically important pathogen. IMPORTANCE Klebsiella pneumoniae is a medically important pathogen that produces a thick protective capsule that is essential for pathogenicity. Phages are natural predators of bacteria, and many encode diverse "capsule depolymerases" which specifically degrade the capsule of their hosts, an exploitable trait for potential therapies. We have determined the first structure of a depolymerase that targets the clinically relevant K2 capsule and have identified its putative active site, providing hints to its mechanism of action. We also show that Klebsiella cells treated with a recombinant form of the depolymerase are stripped of capsule, inhibiting their ability to grow in the presence of serum, demonstrating the anti-infective potential of these robust and readily producible enzymes against encapsulated bacterial pathogens such as K. pneumoniae.
Keywords: Klebsiella; bacteriophages; capsular polysaccharide; cryo-EM; depolymerase.
Figures


References
-
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–E3581. doi: 10.1073/pnas.1501049112. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

