Alveolar macrophages rely on GM-CSF from alveolar epithelial type 2 cells before and after birth
- PMID: 34431978
- PMCID: PMC8404471
- DOI: 10.1084/jem.20210745
Alveolar macrophages rely on GM-CSF from alveolar epithelial type 2 cells before and after birth
Abstract
Programs defining tissue-resident macrophage identity depend on local environmental cues. For alveolar macrophages (AMs), these signals are provided by immune and nonimmune cells and include GM-CSF (CSF2). However, evidence to functionally link components of this intercellular cross talk remains scarce. We thus developed new transgenic mice to profile pulmonary GM-CSF expression, which we detected in both immune cells, including group 2 innate lymphoid cells and γδ T cells, as well as AT2s. AMs were unaffected by constitutive deletion of hematopoietic Csf2 and basophil depletion. Instead, AT2 lineage-specific constitutive and inducible Csf2 deletion revealed the nonredundant function of AT2-derived GM-CSF in instructing AM fate, establishing the postnatal AM compartment, and maintaining AMs in adult lungs. This AT2-AM relationship begins during embryogenesis, where nascent AT2s timely induce GM-CSF expression to support the proliferation and differentiation of fetal monocytes contemporaneously seeding the tissue, and persists into adulthood, when epithelial GM-CSF remains restricted to AT2s.
© 2021 Gschwend et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures





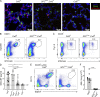
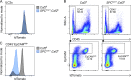

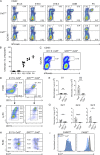



Comment in
-
Alveolar macrophages and epithelial cells: The art of living together.J Exp Med. 2021 Oct 4;218(10):e20211583. doi: 10.1084/jem.20211583. Epub 2021 Sep 7. J Exp Med. 2021. PMID: 34491265 Free PMC article.
References
-
- Basil, M.C., Katzen J., Engler A.E., Guo M., Herriges M.J., Kathiriya J.J., Windmueller R., Ysasi A.B., Zacharias W.J., Chapman H.A., et al. . 2020. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell. 26:482–502. 10.1016/j.stem.2020.03.009 - DOI - PMC - PubMed
-
- Behrens, F., Tak P.P., Østergaard M., Stoilov R., Wiland P., Huizinga T.W., Berenfus V.Y., Vladeva S., Rech J., Rubbert-Roth A., et al. . 2015. MOR103, a human monoclonal antibody to granulocyte-macrophage colony-stimulating factor, in the treatment of patients with moderate rheumatoid arthritis: results of a phase Ib/IIa randomised, double-blind, placebo-controlled, dose-escalation trial. Ann. Rheum. Dis. 74:1058–1064. 10.1136/annrheumdis-2013-204816 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

