Taxonomic Uncertainty and the Anomaly Zone: Phylogenomics Disentangle a Rapid Radiation to Resolve Contentious Species (Gila robusta Complex) in the Colorado River
- PMID: 34432005
- PMCID: PMC8449829
- DOI: 10.1093/gbe/evab200
Taxonomic Uncertainty and the Anomaly Zone: Phylogenomics Disentangle a Rapid Radiation to Resolve Contentious Species (Gila robusta Complex) in the Colorado River
Abstract
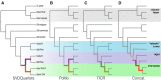
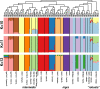
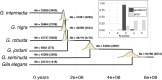
Species are indisputable units for biodiversity conservation, yet their delimitation is fraught with both conceptual and methodological difficulties. A classic example is the taxonomic controversy surrounding the Gila robusta complex in the lower Colorado River of southwestern North America. Nominal species designations were originally defined according to weakly diagnostic morphological differences, but these conflicted with subsequent genetic analyses. Given this ambiguity, the complex was re-defined as a single polytypic unit, with the proposed "threatened" status under the U.S. Endangered Species Act of two elements being withdrawn. Here we re-evaluated the status of the complex by utilizing dense spatial and genomic sampling (n = 387 and >22 k loci), coupled with SNP-based coalescent and polymorphism-aware phylogenetic models. In doing so, we found that all three species were indeed supported as evolutionarily independent lineages, despite widespread phylogenetic discordance. To juxtapose this discrepancy with previous studies, we first categorized those evolutionary mechanisms driving discordance, then tested (and subsequently rejected) prior hypotheses which argued phylogenetic discord in the complex was driven by the hybrid origin of Gila nigra. The inconsistent patterns of diversity we found within G. robusta were instead associated with rapid Plio-Pleistocene drainage evolution, with subsequent divergence within the "anomaly zone" of tree space producing ambiguities that served to confound prior studies. Our results not only support the resurrection of the three species as distinct entities but also offer an empirical example of how phylogenetic discordance can be categorized within other recalcitrant taxa, particularly when variation is primarily partitioned at the species level.
Keywords: RADseq; anomaly zone; conservation genomics; hybridization; phylogenomics; taxonomic uncertainty.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Ahrens D, et al.2016. Rarity and incomplete sampling in DNA-based species delimitation. Syst Biol. 65(3):478–494. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

