Mitogenomes of Accipitriformes and Cathartiformes Were Subjected to Ancestral and Recent Duplications Followed by Gradual Degeneration
- PMID: 34432018
- PMCID: PMC8435663
- DOI: 10.1093/gbe/evab193
Mitogenomes of Accipitriformes and Cathartiformes Were Subjected to Ancestral and Recent Duplications Followed by Gradual Degeneration
Abstract
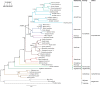
The rearrangement of 37 genes with one control region, firstly identified in Gallus gallus mitogenome, is believed to be ancestral for all Aves. However, mitogenomic sequences obtained in recent years revealed that many avian mitogenomes contain duplicated regions that were omitted in previous genomic versions. Their evolution and mechanism of duplication are still poorly understood. The order of Accipitriformes is especially interesting in this context because its representatives contain a duplicated control region in various stages of degeneration. Therefore, we applied an appropriate PCR strategy to look for duplications within the mitogenomes of the early diverged species Sagittarius serpentarius and Cathartiformes, which is a sister order to Accipitriformes. The analyses revealed the same duplicated gene order in all examined taxa and the common ancestor of these groups. The duplicated regions were subjected to gradual degeneration and homogenization during concerted evolution. The latter process occurred recently in the species of Cathartiformes as well as in the early diverged lineages of Accipitriformes, that is, Sagittarius serpentarius and Pandion haliaetus. However, in other lineages, that is, Pernis ptilorhynchus, as well as representatives of Aegypiinae, Aquilinae, and five related subfamilies of Accipitriformes (Accipitrinae, Circinae, Buteoninae, Haliaeetinae, and Milvinae), the duplications were evolving independently for at least 14-47 Myr. Different portions of control regions in Cathartiformes showed conflicting phylogenetic signals indicating that some sections of these regions were homogenized at a frequency higher than the rate of speciation, whereas others have still evolved separately.
Keywords: Accipitriformes; Aves; Cathartiformes; ancestral state; concerted evolution; control region; duplication; gene order; mitochondrial genome; mitogenome; phylogeny; rearrangement.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures









References
-
- Abbott CL, Double MC, Trueman JW, Robinson A, Cockburn A.. 2005. An unusual source of apparent mitochondrial heteroplasmy: duplicate mitochondrial control regions in Thalassarche albatrosses. Mol Ecol. 14(11):3605–3613. - PubMed
-
- Akiyama T, et al.2017. Gene duplication and concerted evolution of mitochondrial DNA in crane species. Mol Phylogenet Evol. 106:158–163. - PubMed
-
- Anisimova M, Gascuel O.. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 55(4):539–552. - PubMed
-
- Arndt A, Smith MJ.. 1998. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol Biol Evol. 15(8):1009–1016. - PubMed
-
- Bernt M, Braband A, Schierwater B, Stadler PF.. 2013. Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol. 69(2):328–338. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

