Extrinsic interactions in the microenvironment in vivo activate an antiapoptotic multidrug-resistant phenotype in CLL
- PMID: 34432864
- PMCID: PMC8525241
- DOI: 10.1182/bloodadvances.2020003944
Extrinsic interactions in the microenvironment in vivo activate an antiapoptotic multidrug-resistant phenotype in CLL
Abstract
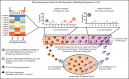
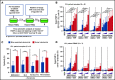
The Bcl-2 inhibitor venetoclax has yielded exceptional clinical responses in chronic lymphocytic leukemia (CLL). However, de novo resistance can result in failure to achieve negative minimal residual disease and predicts poor treatment outcomes. Consequently, additional proapoptotic drugs, such as inhibitors of Mcl-1 and Bcl-xL, are in development. By profiling antiapoptotic proteins using flow cytometry, we find that leukemic B cells that recently emigrated from the lymph node (CD69+/CXCR4Low) in vivo are enriched for cell clusters simultaneously overexpressing multiple antiapoptotic proteins (Mcl-1High/Bcl-xLHigh/Bcl-2High) in both treated and treatment-naive CLL patients. These cells exhibited antiapoptotic resistance to multiple BH-domain antagonists, including inhibitors of Bcl-2, Mcl-1, and Bcl-xL, when tested as single agents in a flow cytometry-based functional assay. Antiapoptotic multidrug resistance declines ex vivo, consistent with resistance being generated in vivo by extrinsic microenvironmental interactions. Surviving "persister" cells in patients undergoing venetoclax treatment are enriched for CLL cells displaying the functional and molecular properties of microenvironmentally induced multidrug resistance. Overcoming this resistance required simultaneous inhibition of multiple antiapoptotic proteins, with potential for unwanted toxicities. Using a drug screen performed using patient peripheral blood mononuclear cells cultured in an ex vivo microenvironment model, we identify novel venetoclax drug combinations that induce selective cytotoxicity in multidrug-resistant CLL cells. Thus, we demonstrate that antiapoptotic multidrug-resistant CLL cells exist in patients de novo and show that these cells persist during proapoptotic treatment, such as venetoclax. We validate clinically actionable approaches to selectively deplete this reservoir in patients.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: C.A.P. receives clinical trial research support from AbbVie, via the UVA Office of Sponsored Programs. C.A.P. has received clinical trial research support from Genentech/Roche, TG Therapeutics, Infinity, BeiGene, and Acerta/AstraZeneca. C.A.P. has also done consulted for Genentech, Bayer, BeiGene, Janssen, and Pharmacyclics. M.E.W. has received clinical trial research support from Janssen, Pharmacyclics. and TG Therapeutics and consulting fees from Gilead Sciences, AbbVie, Astra-Zeneca, Celgene, Janssen, Verastem, and TG Therapeutics. The remaining authors declare no competing financial interests.
Figures




References
-
- Souers AJ, Leverson JD, Boghaert ER, et al. . ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19(2):202-208. - PubMed
-
- Thompson M, Brander D, Nabhan C, Mato A.. minimal residual disease in chronic lymphocytic leukemia in the era of novel agents: a review. JAMA Oncol. 2018;4(3):394-400. - PubMed
-
- Molica S, Giannarelli D, Montserrat E.. Minimal residual disease and survival outcomes in patients with chronic lymphocytic leukemia: a systematic review and meta-analysis. Clin Lymphoma Myeloma Leuk. 2019;19(7):423-430. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

