Global spread of Salmonella Enteritidis via centralized sourcing and international trade of poultry breeding stocks
- PMID: 34433807
- PMCID: PMC8387372
- DOI: 10.1038/s41467-021-25319-7
Global spread of Salmonella Enteritidis via centralized sourcing and international trade of poultry breeding stocks
Abstract
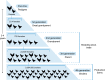
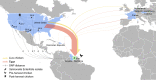
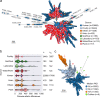
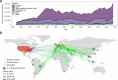
A pandemic of Salmonella enterica serotype Enteritidis emerged in the 1980s due to contaminated poultry products. How Salmonella Enteritidis rapidly swept through continents remains a historical puzzle as the pathogen continues to cause outbreaks and poultry supply becomes globalized. We hypothesize that international trade of infected breeding stocks causes global spread of the pathogen. By integrating over 30,000 Salmonella Enteritidis genomes from 98 countries during 1949-2020 and international trade of live poultry from the 1980s to the late 2010s, we present multifaceted evidence that converges on a high likelihood, global scale, and extended protraction of Salmonella Enteritidis dissemination via centralized sourcing and international trade of breeding stocks. We discovered recent, genetically near-identical isolates from domestically raised poultry in North and South America. We obtained phylodynamic characteristics of global Salmonella Enteritidis populations that lend spatiotemporal support for its dispersal from centralized origins during the pandemic. We identified concordant patterns of international trade of breeding stocks and quantitatively established a driving role of the trade in the geographic dispersal of Salmonella Enteritidis, suggesting that the centralized origins were infected breeding stocks. Here we demonstrate the value of integrative and hypothesis-driven data mining in unravelling otherwise difficult-to-probe pathogen dissemination from hidden origins.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Shahbandeh, M. Production of meat worldwide from 2016 to 2020, by type (in million metric tons). Statistahttps://www.statista.com/statistics/237632/production-of-meat-worldwide-... (2020). Accessed June 09, 2021.
-
- Food and Agriculture Organization of the United Nations. Food and agriculture data http://www.fao.org/faostat/en/#home (Food and Agriculture Organization of the United Nations, 2020). Accessed June 09, 2021.
-
- Mottet A, Tempio G. Global poultry production: current state and future outlook and challenges. World’s Poult. Sci. J. 2017;73:245–256. doi: 10.1017/S0043933917000071. - DOI
-
- Shrader HL. The Chicken-of-Tomorrom Program: its influence on “meat-type” poultry production. Poult. Sci. 1952;31:3–10. doi: 10.3382/ps.0310003. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

