Benchmarking laboratory processes to characterise low-biomass respiratory microbiota
- PMID: 34433845
- PMCID: PMC8387476
- DOI: 10.1038/s41598-021-96556-5
Benchmarking laboratory processes to characterise low-biomass respiratory microbiota
Abstract
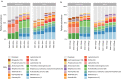
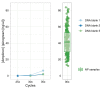
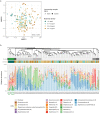
The low biomass of respiratory samples makes it difficult to accurately characterise the microbial community composition. PCR conditions and contaminating microbial DNA can alter the biological profile. The objective of this study was to benchmark the currently available laboratory protocols to accurately analyse the microbial community of low biomass samples. To study the effect of PCR conditions on the microbial community profile, we amplified the 16S rRNA gene of respiratory samples using various bacterial loads and different number of PCR cycles. Libraries were purified by gel electrophoresis or AMPure XP and sequenced by V2 or V3 MiSeq reagent kits by Illumina sequencing. The positive control was diluted in different solvents. PCR conditions had no significant influence on the microbial community profile of low biomass samples. Purification methods and MiSeq reagent kits provided nearly similar microbiota profiles (paired Bray-Curtis dissimilarity median: 0.03 and 0.05, respectively). While profiles of positive controls were significantly influenced by the type of dilution solvent, the theoretical profile of the Zymo mock was most accurately analysed when the Zymo mock was diluted in elution buffer (difference compared to the theoretical Zymo mock: 21.6% for elution buffer, 29.2% for Milli-Q, and 79.6% for DNA/RNA shield). Microbiota profiles of DNA blanks formed a distinct cluster compared to low biomass samples, demonstrating that low biomass samples can accurately be distinguished from DNA blanks. In summary, to accurately characterise the microbial community composition we recommend 1. amplification of the obtained microbial DNA with 30 PCR cycles, 2. purifying amplicon pools by two consecutive AMPure XP steps and 3. sequence the pooled amplicons by V3 MiSeq reagent kit. The benchmarked standardized laboratory workflow presented here ensures comparability of results within and between low biomass microbiome studies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

