Potential Therapeutic Role of HDAC Inhibitors in FUS-ALS
- PMID: 34434087
- PMCID: PMC8380926
- DOI: 10.3389/fnmol.2021.686995
Potential Therapeutic Role of HDAC Inhibitors in FUS-ALS
Abstract
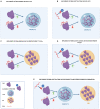
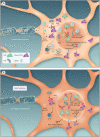
Mutations in the FUS gene cause amyotrophic lateral sclerosis (ALS-FUS). However, the exact pathogenic mechanism of mutant fused in sarcoma (FUS) protein is not completely understood. FUS is an RNA binding protein (RBP) localized predominantly in the nucleus, but ALS-linked FUS mutations can affect its nuclear localization signal impairing its import into the nucleus. This mislocalization to the cytoplasm facilitates FUS aggregation in cytoplasmic inclusions. Therapies targeting post translational modifications are rising as new treatments for ALS, in particular acetylation which could have a role in the dynamics of RBPs. Research using histone deacetylase (HDAC) inhibitors in FUS-ALS models showed that HDACs can influence cytoplasmic FUS localization. Inhibition of HDACs could promote acetylation of the FUS RNA binding domain (RRM) and altering its RNA interactions resulting in FUS maintenance in the nucleus. In addition, acetylation of FUS RRMs might also favor or disfavor its incorporation into pathological inclusions. In this review, we summarize and discuss the evidence for the potential role of HDACs in the context of FUS-ALS and we propose a new hypothesis based on this overview.
Keywords: ALS; FUS; HDAC inhibitors; LLPS; RNA; RRM; acetylation; mislocalization.
Copyright © 2021 Tejido, Pakravan and Bosch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Andersson M. K., Ståhlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., et al. . (2008). The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 9, 9–37. 10.1186/1471-2121-9-37 - DOI - PMC - PubMed
-
- Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., et al. . (2006). TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611. 10.1016/j.bbrc.2006.10.093 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

