Genome-wide analysis of the lignin toolbox for morus and the roles of lignin related genes in response to zinc stress
- PMID: 34434666
- PMCID: PMC8351576
- DOI: 10.7717/peerj.11964
Genome-wide analysis of the lignin toolbox for morus and the roles of lignin related genes in response to zinc stress
Abstract
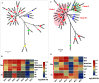
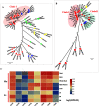
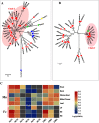
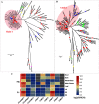
Mulberry (Morus, Moraceae) is an important economic plant with nutritional, medicinal, and ecological values. Lignin in mulberry can affect the quality of forage and the saccharification efficiency of mulberry twigs. The availability of the Morus notabilis genome makes it possible to perform a systematic analysis of the genes encoding the 11 protein families specific to the lignin branch of the phenylpropanoid pathway, providing the core genes for the lignin toolbox in mulberry. We performed genome-wide screening, which was combined with de novo transcriptome data for Morus notabilis and Morus alba variety Fengchi, to identify putative members of the lignin gene families followed by phylogenetic and expression profile analyses. We focused on bona fide clade genes and their response to zinc stress were further distinguished based on expression profiles using RNA-seq and RT-qPCR. We finally identified 31 bona fide genes in Morus notabilis and 25 bona fide genes in Fengchi. The putative function of these bona fide genes was proposed, and a lignin toolbox that comprised 19 genes in mulberry was provided, which will be convenient for researchers to explore and modify the monolignol biosynthesis pathway in mulberry. We also observed changes in the expression of some of these lignin biosynthetic genes in response to stress caused by excess zinc in Fengchi and proposed that the enhanced lignin biosynthesis in lignified organs and inhibition of lignin biosynthesis in leaf is an important response to zinc stress in mulberry.
Keywords: Gene family; Genome-wide; Lignin; Mulberry; Zinc stress.
© 2021 Chao et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou JP, Ullmann P, Pauly M, Lapierre C, Werck-Reichhart D. A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiology. 2006;140(1):30–48. doi: 10.1104/pp.105.069690. - DOI - PMC - PubMed
-
- Bhardwaj R, Handa N, Sharma R, Kaur H, Kohli S, Kumar V, Kaur P. Lignins and abiotic stress: an overview. In: Ahmad P, Wani M, editors. Physiological Mechanisms and Adaptation Strategies in Plants Under Changing Environment. New York: Springer; 2014. pp. 267–296.
LinkOut - more resources
Full Text Sources
Miscellaneous

