Phylum barrier and Escherichia coli intra-species phylogeny drive the acquisition of antibiotic-resistance genes
- PMID: 34435947
- PMCID: PMC8549366
- DOI: 10.1099/mgen.0.000489
Phylum barrier and Escherichia coli intra-species phylogeny drive the acquisition of antibiotic-resistance genes
Abstract
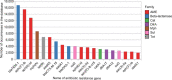
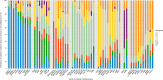
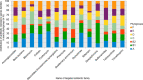
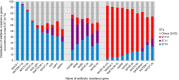
Escherichia coli is a ubiquitous bacterium that has been widely exposed to antibiotics over the last 70 years. It has adapted by acquiring different antibiotic-resistance genes (ARGs), the census of which we aim to characterize here. To do so, we analysed 70 301 E. coli genomes obtained from the EnteroBase database and detected 1 027 651 ARGs using the AMRFinder, Mustard and ResfinderFG ARG databases. We observed a strong phylogroup and clonal lineage specific distribution of some ARGs, supporting the argument for epistasis between ARGs and the strain genetic background. However, each phylogroup had ARGs conferring a similar antibiotic class resistance pattern, indicating phenotypic adaptive convergence. The G+C content or the type of ARG was not associated with the frequency of the ARG in the database. In addition, we identified ARGs from anaerobic, non-Proteobacteria bacteria in four genomes of E. coli, supporting the hypothesis that the transfer between anaerobic bacteria and E. coli can spontaneously occur but remains exceptional. In conclusion, we showed that phylum barrier and intra-species phylogenetic history are major drivers of the acquisition of a resistome in E. coli.
Keywords: Escherichia coli; antibiotic-resistance genes.
Conflict of interest statement
The authors declare that there are no conflicts of interest
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

