A human apolipoprotein L with detergent-like activity kills intracellular pathogens
- PMID: 34437126
- PMCID: PMC8422858
- DOI: 10.1126/science.abf8113
A human apolipoprotein L with detergent-like activity kills intracellular pathogens
Abstract
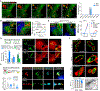
Activation of cell-autonomous defense by the immune cytokine interferon-γ (IFN-γ) is critical to the control of life-threatening infections in humans. IFN-γ induces the expression of hundreds of host proteins in all nucleated cells and tissues, yet many of these proteins remain uncharacterized. We screened 19,050 human genes by CRISPR-Cas9 mutagenesis and identified IFN-γ-induced apolipoprotein L3 (APOL3) as a potent bactericidal agent protecting multiple non-immune barrier cell types against infection. Canonical apolipoproteins typically solubilize mammalian lipids for extracellular transport; APOL3 instead targeted cytosol-invasive bacteria to dissolve their anionic membranes into human-bacterial lipoprotein nanodiscs detected by native mass spectrometry and visualized by single-particle cryo-electron microscopy. Thus, humans have harnessed the detergent-like properties of extracellular apolipoproteins to fashion an intracellular lysin, thereby endowing resident nonimmune cells with a mechanism to achieve sterilizing immunity.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




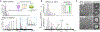
Comment in
-
Rethinking immunology.Science. 2021 Jul 16;373(6552):276-277. doi: 10.1126/science.abj5637. Science. 2021. PMID: 34437138 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

