Recognition of Divergent Viral Substrates by the SARS-CoV-2 Main Protease
- PMID: 34437808
- PMCID: PMC8424689
- DOI: 10.1021/acsinfecdis.1c00237
Recognition of Divergent Viral Substrates by the SARS-CoV-2 Main Protease
Abstract
The main protease (Mpro) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease (COVID-19), is an ideal target for pharmaceutical inhibition. Mpro is conserved among coronaviruses and distinct from human proteases. Viral replication depends on the cleavage of the viral polyprotein at multiple sites. We present crystal structures of SARS-CoV-2 Mpro bound to two viral substrate peptides. The structures show how Mpro recognizes distinct substrates and how subtle changes in substrate accommodation can drive large changes in catalytic efficiency. One peptide, constituting the junction between viral nonstructural proteins 8 and 9 (nsp8/9), has P1' and P2' residues that are unique among the SARS-CoV-2 Mpro cleavage sites but conserved among homologous junctions in coronaviruses. Mpro cleaves nsp8/9 inefficiently, and amino acid substitutions at P1' or P2' can enhance catalysis. Visualization of Mpro with intact substrates provides new templates for antiviral drug design and suggests that the coronavirus lifecycle selects for finely tuned substrate-dependent catalytic parameters.
Keywords: Mpro; SARS-CoV-2; protease; virology.
Conflict of interest statement
Mark Namchuk is the recipient of funding from AbbVie. The funding did not contribute to this work.
Figures

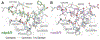
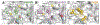
References
-
- Anand K; Ziebuhr J; Wadhwani P; Mesters JR; Hilgenfeld R, Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

