Bottom-up proteomic analysis of human adult cardiac tissue and isolated cardiomyocytes
- PMID: 34437879
- PMCID: PMC9620472
- DOI: 10.1016/j.yjmcc.2021.08.008
Bottom-up proteomic analysis of human adult cardiac tissue and isolated cardiomyocytes
Abstract
The heart is composed of multiple cell types, each with a specific function. Cell-type-specific approaches are necessary for defining the intricate molecular mechanisms underlying cardiac development, homeostasis, and pathology. While single-cell RNA-seq studies are beginning to define the chamber-specific cellular composition of the heart, our views of the proteome are more limited because most proteomics studies have utilized homogenized human cardiac tissue. To promote future cell-type specific analyses of the human heart, we describe the first method for cardiomyocyte isolation from cryopreserved human cardiac tissue followed by flow cytometry for purity assessment. We also describe a facile method for preparing isolated cardiomyocytes and whole cardiac tissue homogenate for bottom-up proteomic analyses. Prior experience in dissociating cardiac tissue or proteomics is not required to execute these methods. We compare different sample preparation workflows and analysis methods to demonstrate how these can impact the depth of proteome coverage achieved. We expect this how-to guide will serve as a starting point for investigators interested in general and cell-type-specific views of the cardiac proteome.
Keywords: Cardiomyocyte isolation; Data independent acquisition; Proteomics; S-trap.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest
R.L.G. is on the advisory board of ProtiFi, LLC and receives no compensation of any kind for this role.
Figures

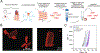




References
-
- Fountoulaki K, Cardiothoracic Intensive Care Unit, Onassis Cardiac Surgery Centre, Athens, Greece, N. Dagres, Second University Department of Cardiology, Attikon General Hospial, University of Athens, Athens, Greece, Cellular communications in the heart, Card. Fail. Rev 1 (2015) 64, 10.15420/cfr.2015.1.2.64. - DOI - PMC - PubMed
-
- Asp M, Giacomello S, Larsson L, Wu C, Fürth D, Qian X, Wärdell E, Custodio J, Reimegård J, Salmén F, Österholm C, Ståhl PL, Sundström E, Åkesson E, Bergmann O, Bienko M, Månsson-Broberg A, Nilsson M, Sylvén C, Lundeberg J, A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart, Cell 179 (2019) 1647–1660.e19, 10.1016/j.cell.2019.11.025. - DOI - PubMed
-
- Tucker Nathan R., Mark Chaffin, Stephen J. Fleming, Amelia W. Hall, Victoria A. Parsons, Kenneth C. Bedi, Amer-Denis Akkad, Caroline N. Herndon, Alessandro Arduini, Irinna Papangeli, Carolina Roselli, François Aguet, Hoan Choi Seung, Kristin G. Ardlie, Mehrtash Babadi, Kenneth B. Margulies, Christian M. Stegmann, Patrick T. Ellinor, Transcriptional and cellular diversity of the human heart, Circulation 142 (2020) 466–482, 10.1161/CIRCULATIONAHA.119.045401. - DOI - PMC - PubMed
-
- Litviñuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, Nadelmann ER, Roberts K, Tuck L, Fasouli ES, DeLaughter DM, McDonough B, Wakimoto H, Gorham JL, Samari S, Mahbubani KT, Saeb-Parsy K, Patone G, Boyle JJ, Zhang H, Zhang H, Viveiros A, Oudit GY, Bayraktar OA, Seidman JG, Seidman CE, Noseda M, Hubner N, Teichmann SA, Cells of the adult human heart, Nature 588 (2020) 466–472, 10.1038/s41586-020-2797-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

