Multicenter DSC-MRI-Based Radiomics Predict IDH Mutation in Gliomas
- PMID: 34439118
- PMCID: PMC8391559
- DOI: 10.3390/cancers13163965
Multicenter DSC-MRI-Based Radiomics Predict IDH Mutation in Gliomas
Abstract
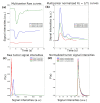
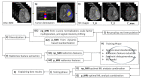
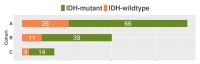
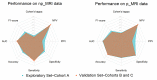
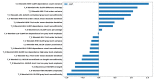
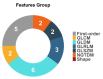
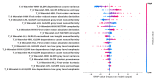
To address the current lack of dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI)-based radiomics to predict isocitrate dehydrogenase (IDH) mutations in gliomas, we present a multicenter study that featured an independent exploratory set for radiomics model development and external validation using two independent cohorts. The maximum performance of the IDH mutation status prediction on the validation set had an accuracy of 0.544 (Cohen's kappa: 0.145, F1-score: 0.415, area under the curve-AUC: 0.639, sensitivity: 0.733, specificity: 0.491), which significantly improved to an accuracy of 0.706 (Cohen's kappa: 0.282, F1-score: 0.474, AUC: 0.667, sensitivity: 0.6, specificity: 0.736) when dynamic-based standardization of the images was performed prior to the radiomics. Model explainability using local interpretable model-agnostic explanations (LIME) and Shapley additive explanations (SHAP) revealed potential intuitive correlations between the IDH-wildtype increased heterogeneity and the texture complexity. These results strengthened our hypothesis that DSC-MRI radiogenomics in gliomas hold the potential to provide increased predictive performance from models that generalize well and provide understandable patterns between IDH mutation status and the extracted features toward enabling the clinical translation of radiogenomics in neuro-oncology.
Keywords: IDH mutation; dynamic susceptibility contrast MRI; explainability; external validation; generalizability; gliomas; radiomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
-
- Lu V.M., O’Connor K.P., Shah A.H., Eichberg D.G., Luther E.M., Komotar R.J., Ivan M.E. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: A systematic review of the contemporary literature. J. Neurooncol. 2020;148:221–229. doi: 10.1007/s11060-020-03528-2. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

