Discovering the Protective Effects of Resveratrol on Aflatoxin B1-Induced Toxicity: A Whole Transcriptomic Study in a Bovine Hepatocyte Cell Line
- PMID: 34439473
- PMCID: PMC8388899
- DOI: 10.3390/antiox10081225
Discovering the Protective Effects of Resveratrol on Aflatoxin B1-Induced Toxicity: A Whole Transcriptomic Study in a Bovine Hepatocyte Cell Line
Abstract
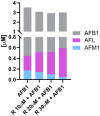
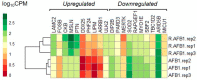
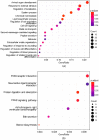
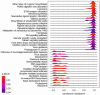
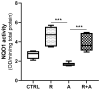
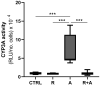
Aflatoxin B1 (AFB1) is a natural feed and food contaminant classified as a group I carcinogen for humans. In the dairy industry, AFB1 and its derivative, AFM1, are of concern for the related economic losses and their possible presence in milk and dairy food products. Among its toxic effects, AFB1 can cause oxidative stress. Thus, dietary supplementation with natural antioxidants has been considered among the strategies to mitigate AFB1 presence and its toxicity. Here, the protective role of resveratrol (R) has been investigated in a foetal bovine hepatocyte cell line (BFH12) exposed to AFB1, by measuring cytotoxicity, transcriptional changes (RNA sequencing), and targeted post-transcriptional modifications (lipid peroxidation, NQO1 and CYP3A enzymatic activity). Resveratrol reversed the AFB1-dependent cytotoxicity. As for gene expression, when administered alone, R induced neglectable changes in BFH12 cells. Conversely, when comparing AFB1-exposed cells with those co-incubated with R+AFB1, greater transcriptional variations were observed (i.e., 840 DEGs). Functional analyses revealed that several significant genes were involved in lipid biosynthesis, response to external stimulus, drug metabolism, and inflammatory response. As for NQO1 and CYP3A activities and lipid peroxidation, R significantly reverted variations induced by AFB1, mostly corroborating and/or completing transcriptional data. Outcomes of the present study provide new knowledge about key molecular mechanisms involved in R antioxidant-mediated protection against AFB1 toxicity.
Keywords: RNAseq; aflatoxicosis; aflatoxin B1; antioxidants; cancer; cattle; liver; mycotoxins; resveratrol; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Monbaliu S., Van Poucke C., Detavernier C., Dumoulin F., Van De Velde M., Schoeters E., Van Dyck S., Averkieva O., Van Peteghem C., De Saeger S. Occurrence of Mycotoxins in Feed as Analyzed by a Multi-Mycotoxin LC-MS/MS Method. J. Agric. Food Chem. 2010;58:66–71. doi: 10.1021/jf903859z. - DOI - PubMed
-
- Shi F., Seng X., Tang H., Zhao S., Deng Y., Jin R., Li Y. Effect of Low Levels of Aflatoxin B1 on Performance, Serum Biochemistry, Hepatocyte Apoptosis and Liver Histopathological Changes of Cherry Valley Ducks. J. Anim. Vet. Adv. 2013;12:1126–1130. doi: 10.3923/javaa.2013.1126.1130. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

